Abstract
Polymer electrolyte membrane (PEM) technologies hold promise for sustainable energy solutions, yet pinhole-related challenges persist. Our research introduces a novel biohybrid approach to self-healing, enhancing multiple healing cycles with minimal membrane disruption. Initial steps involve immobilizing enzymes on a polymeric membrane. This study establishes the immobilization process and analytical framework through enzyme immobilization on polypropylene. Applicability and stability are investigated, laying groundwork for potential Nafion™ applications and advancing climate neutral energy.
Qualitative analysis employs colorimetric p-NPA assay on polypropylene-immobilized lipase from Candida rugosa (CRL) and Lipase B from Candida antarctica (CALB). Both enzymes hold their temperature optimum at 50°C which is increased by 10°C via immobilization. Diisopropylcarbodiimide (DIC) is optimal for immobilization. Synchronous enzyme and DIC addition is advantageous. After 8 reuse cycles, immobilized enzymes retain 54.3% residual activity. Immobilizates exposed to PEM fuel cell conditions show better stability due to covalent immobilization than free CRL. Yet, declines occur under stressors like 60 °C and concentrated alcohol. Immobilizates remain resilient at pH 3 and under oxidizing as well as reducing conditions constituted by varied gas atmospheres. Considering PEM fuel cells' operational range, in-depth investigations across conditions are vital. Future studies target long-term PEM fuel cell lifespans, focusing on extremophilic enzymes or modifications for high-temperature stability. Subsequently, the transferability of the immobilization method to Nafion™ shall be deliberated based on the outcomes.
Author Contributions
Academic Editor: Mezni Ali, Department of Life Sciences, University of Carthage
Checked for plagiarism: Yes
Review by: Single-blind
Copyright © 2023 Patrizia Gartner, et al.
 This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Competing interests
The authors have declared that no competing interests exist.
Citation:
Introduction
Polymer electrolyte membrane (PEM) fuel cells and electrolyzers are highly important technologies that are being extensively pursued for greener mobility and energy supply. These technologies share a common core component – the PEM. However, these PEMs are prone to pinholes, a wear phenomenon that leads to performance degradation and early application failure 1, 2, 3. Thus, the overarching objective of this research is to develop a self-healing mechanism for pinholes within PEM fuel cell and electrolyzer membranes.
Previous investigations into self-healing mechanisms for PEM fuel cells have explored techniques such as integrating additional materials or fillers that melt or encapsulate, releasing when cracks occur 4, 5. These additives are embedded within the membrane and used up after a self-healing event, restricting the process to a single-use at a specific site. Furthermore, such modifications have been reported to negatively impact fuel cell performance.
In contrast, our approach employs a biohybrid system, utilizing filler-generating enzymes immobilized directly onto the membrane's surface. These enzymes facilitate filler polymerization from externally supplied monomers, effectively sealing cracks. This technique allows for repeated self-healing at the same location, thereby mitigating the negative impact on PEM fuel cell performance in theory 6.
Initiating this approach involves enzyme immobilization on the membrane's surface to enable targeted filler production. Enzyme immobilization enables multiple self-healing cycles and enhances enzyme stability towards solvents and higher temperatures, which is required in the fuel cell 7, 8. Nafion™, a costly fluorinated polymer by DuPont, is commonly used in PEM applications. To establish the immobilization method and analytical framework, we began with cost-effective polypropylene membranes as model substance, given their manageable characteristics. Comparable to Nafion™, polypropylene can be functionalized for enzyme immobilization.
Various enzyme immobilization methods have been explored in literature, encompassing entrapment, adsorption, cross-linking, affinity, and covalent bonding 9. Covalent bonding necessitates functional groups, which polypropylene lacks. To overcome this, pre-immobilization surface oxidation introduces anchoring points. Key challenges in enzyme immobilization for PEM fuel cells, inspired by Luo et al.9, include reconciling enzyme immobilization strategies with application and membrane-specific requirements. Enzyme survival under fuel cell conditions and stability are crucial factors, favoring covalent immobilization over non-covalent methods, despite reduced activity. Modifying membrane treatment might affect membrane stability, and substrate accessibility hinges on the fuel cell system’s pore sizes, physicochemical interactions, and flow rate. Balancing enzyme loading and membrane function is crucial, as higher enzyme loading could impede proton transport. Potential system fouling and the transferability of the here established immobilization strategy to Nafion™ are also pertinent considerations.
Our study establishes covalent enzyme immobilization of Candida rugosa (CRL) and Candida antarctica (CALB) lipases on polypropylene membranes. We assess their applicability in PEM fuel cells by analyzing immobilization stability against fuel cell conditions. Storage, temperature, pH, gas atmospheres, solvent effects, and multiple self-healing capabilities are investigated. Further discussions center on the transferability of this strategy to Nafion™ immobilization and answer the question if the strategy can serve as a role model.
Material and Methods
Enzyme solutions utilized lipase from Candida rugosa (CRL, 5.2 U/mg, Sigma-Aldrich) and lipase B from Candida antarctica (CALB, recomb. Asp. Oryzae, Sigma-Aldrich). Stock solution contained 2 g/L in 50 mM phosphate buffer pH 7.2 (composed of 4.25 g sodium chloride, 1.1 g disodium hydrogen phosphate, 2.6 g sodium dihydrogen phosphate dihydrate, 500 mL ultrapure water), mixed on a rotator at 30 rpm for 1 h. Concentrations are based on purchased enzyme formulation mass, not pure enzyme mass. Enzyme activity measured using para-nitrophenol acetate assay (p-NPA). For free lipases, stock solution diluted to 0.1 g/L. Lipase solutions (495 µL) were added in triplicate to microreaction tubes (Eppendorf), using 495 µL phosphate buffer as blank. For polypropylene (PP)-immobilized lipases, membranes in microreaction tubes with 495 µL phosphate buffer, fully covered. Substrate solution (5 mM para-nitrophenol acetate (S. Jansen) in ethanol) added (5 µL), vortex mixed (5 s), incubated (20 min, 1000 rpm, 23°C), mixed (5 s). 200 µL of triplicates pipetted into 96-well microtiter plate (PS, F-bottom, clear, Greiner Bio-One) and absorption measured at 410 nm immediately using microplate photometer (Infinite M200 pro, Tecan Group).
Immobilization of CRL and CALB to a functionalized PP-membrane (model 4000-00-03-V, Leitz) was executed using the schema after Hermanson, see Figure 1.
Figure 1.Functionalization of methyl groups to carboxyl groups on polypropylene via potassium dichromate. Modified after Hermanson 10.
Methyl groups on PP-membranes were oxidized to carboxyl groups using potassium dichromate. Membranes of a size of 13 x 80 mm were immersed in 30 mL acetone for 10 min at 23°C and then in 15 mL oxidation solution (3 g potassium dichromate, 11.4 mL ultrapure water, 9.58 mL 96% sulfuric acid) for 10 min at 30 °C, 500 rpm. After 3 ultrapure water washes, enzymes were covalently immobilized on functionalized PP-membranes using carbodiimides as carboxyl group activators. Each membrane received 15 mL phosphate buffer (50 mM, pH 7.2) with 1 g/L enzyme and 3.221 mmol/L opylcarbodiimide (DIC, Thermo Fisher Scientific). Incubation was 18 h at 4 °C, 5 rpm, followed by 30-min washing at 23°C, 500 rpm. Membranes with a diameter of 6 mm were then punched out using punch pliers. Immobilization success was determined by p-NPA assay triplicate measurements.
For optimiziation experiments instead of DIC, 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC, Carbosynth Holdings Limited) and dicyclohexylcarbodiimide (DCC, Carl Roth) were tested. In addition, an immobilizate was prepared in which the lipase was adsorptively immobilized onto the polypropylene membrane without the addition of any carbodiimide. Further, immobilizates of CRL were prepared in 3 different ways. Method A: PP-membranes were incubated in a plastic tube for 60 min at 30 °C and 500 rpm with 15 mL of phosphate buffer containing 3.221 mmol/L DIC, then transferred and washed for 5 min in a fresh plastic tube, after which 15 mL of a solution of phosphate buffer containing 2 g/L enzyme was added. Method B: Analogous to method A only without washing. Method C: As described in section 2.2. All three plastic tubes were then incubated for 18 h at 4 °C at 5 rpm.
For reusability experiments, immobilizates were prepared and an initial activity assay was performed. Then, the membranes were transferred to an Eppendorf reaction vessel containing 500 μL phosphate buffer and washed for 5 s on the vortex at 23 °C. Subsequently, the activity of the same membranes was determined using a second p-NPA assay. These steps were repeated 8 times.
The enzyme’s stability in terms of storage, temperature, acidic pH, alcoholic media, and gas atmospheres, were investigated using a solution of 2 g/L CRL in phosphate buffer, pH 7.2 as well as PP-immobilizates of CRL.
Storage stability conditions were 23 °C and protected from light. 15 free CRL samples and 15 immobilized samples were stored for various times 0, 14, and 28 d, and enzyme activity was determined using a standard p-NPA assay.
To determine temperature stability, free lipase solutions and immobilizates were exposed for 3 d at pH 7.2 and 60 °C or 4 °C, and enzyme activity was measured via a standard assay at room temperature.
To assess stability against acidic pH, free CRL solution and immobilizates were stored for 3 days at 4°C in 50 mM citrate buffer at pH 3 (composed of 138 mg tri-sodium citrate dihydrate, 870 mg citric acid, 100 mL ultrapure water). Immobilization activities were pre- and post-measured using a p-NPA assay. Free CRL solutions underwent rebuffering by introducing solutions into Slide-A-Lyzer™ MINI dialysis units (Thermo Scientific), followed by enzyme activity measurement. Loss of enzyme activity due to dialysis was quantified by comparing substrate turnover of stored, non-rebuffered CRL (56.8 %) with dialyzed (48.3 %) counterparts, resulting in a 14.9 % activity loss due to dialysis treatment.
Enzyme stability of free CRL solutions on alcoholic components was tested by preparing 2g/L enzyme solution in phosphate buffer with 5 w/w% ethanol, resp. 60 w/w% 1-propanol, and resp. a combination of both. 500 µL of each enzyme solution was added to a Slide-A-Lyzer™ MINI dialysis units for rebuffering. The immobilized samples were incubated together with 500 μL of the respective alcohol solutions in an Eppendorf reaction vessel at 23 °C for 5 min at 1000 rpm, rinsed with 500 μL ultrapure water for 5 s on the vortex, and an assay was performed.
To assess enzyme stability under different gas atmospheres, 50 mL free lipase solutions resp. 3 spiked PP-immobilized lipases were sealed in 250 mL serum bottles. Blank samples with phosphate buffer pH 7.2 were included. Cycles of negative (0.1 bar) and positive pressures (0.7 / 0.9 bar) using nitrogen (Air Liquide) were repeated 20 times to replace dissolved gases with inert nitrogen. Oxygen resp. hydrogen (Air Liquide) was added to the vacuumed septum bottles and brought to ambient pressure by inserting a cannula. Incubation was at 13 °C, 250 rpm, and enzyme activity was determined using p-NPA assay after 3 and 5 days. Buffer solutions incubated together with the samples were used as blanks to account for possible influences of gas atmospheres on the autohydrolysis of p-NPA.
Results and Discussion
Functionalization and immobilization of enzymes on a polypropylene membrane
For qualitative analysis, we conducted a colorimetric p-NPA assay on Lipase from Candida rugosa (CRL) and Lipase B from Candida antarctica (CALB) immobilized on polypropylene. Substrate conversion versus temperature was measured (Figure 2). CRL immobilizate displayed 2 to 3-fold higher activity across temperatures due to differing enzyme preparations. CRL preparation had 18.4% protein content, CALB had 7.5%. Composition confirmed by SDS-PAGE. At 20 °C, immobilized CRL showed significantly higher substrate conversion (15.3%) than CALB (3.3%). This discrepancy underscores the suitability of the standard assay conducted at 23 °C, which appears more apt for CRL than for CALB. Both immobilizates peaked at 50 °C, with CRL's optimum elevated from 40 °C (Figure 3).
Figure 2.Correlation of p-NPA substrate conversion over temperature of the immobilizate of lipase from Candida rugosa (CRL) on polypropylene on the left and Lipase B from Candida antarctica (CALB) on polypropylene on the right at different temperatures. The immobilizates were prepared with 2 g/L enzyme formulation. Assay duration: 20 min. Both immobilizates have a temperature optimum at 50 °C.
Figure 3.The immobilization of lipase from Candida rugosa (CRL) led to an increase of Topt by 10°C.
The optimization of the immobilization process involved testing various carbodiimides (EDC, DIC, and DCC) and different application strategies, as illustrated in Figure 4. DIC emerged as the most promising option for the immobilization method, with a rate of 14.6%. Notably, DCC exhibited limited solubility in the enzyme solution and did not contribute significantly to the formation of peptide bonds between the enzymes and carboxyl groups on the functionalized polypropylene membrane. Despite the anticipated superior immobilization potential of EDC, its water solubility did not translate into markedly improved results. The insolubility of DIC in water, as noted in the literature, was not evident, indicating its favorable behavior. This divergence might be attributed to incomplete oxidation of the membrane's hydrophobic methyl groups to hydrophilic carboxyl groups during functionalization, which could enable better action of DIC on the membrane compared to EDC.
Figure 4.p-NPA substrate conversion of the immobilizates of lipase from Candida rugosa (CRL) on polypropylene. Left: prepared with different carbodiimides EDC, DIC, DCC; Ads: Adsorptive. Right: prepared by different methods. A: Preactivation by DIC followed by washing; B: Preactivation by DIC without subsequent washing; C: Simultaneous addition of DIC and enzyme. The use of DIC and method C show the highest substrate conversion.
Several aspects of the immobilization process were explored, including the sequence of carbodiimide addition and the impact of washing steps between process stages. Optimal results were achieved using DIC and method C, yielding an immobilization rate of 19.6%. The influence of multiple washing steps following the addition of DIC did not alter subsequent immobilization (A vs. B), suggesting that adsorptive immobilization was unlikely to be reduced. Furthermore, simultaneous enzyme and DIC addition (method C) proved more advantageous than preactivation by DIC (methods A and B). This difference might be attributed to DIC's activation of carboxyl groups on both the membrane and enzyme surfaces, potentially enabling enzyme crosslinking and increasing the overall number of immobilized enzymes. An advantage of crosslinking would be that more enzymes can participate in the future polymerization reactions, but activity could also be reduced by steric hindrance.
The study also examined the reusability of immobilized enzymes after eight activity cycles (Figure 5). A typical initial activity loss of 14 % was observed between the first and second cycles. Impressively, after eight cycles, the immobilized enzymes retained over 54 % residual activity, indicating the potential for multiple enzymatic self-healing cycles within a PEM fuel cell. To elucidate the source of the activity loss, a negative control was implemented. Immobilizates were transferred equivalently between reaction vessels to simulate washing during eight activity assays. The activity of the immobilized enzymes did not decrease despite multiple washings, thus confirming that denaturation of the immobilized enzymes was the primary cause of the observed activity loss.
Figure 5.Relative activity of the immobilizates of lipase from Candida rugosa (CRL) on polypropylene over the number of activity cycles. Immobilizates are prepared after method C. Storage at: 23 °C, pH 7.2, in PBS buffer. After an initial moderate loss of activity with around 10 % decrease per cycle, only marginal reductions in activity are seen from the fifth reuse onwards.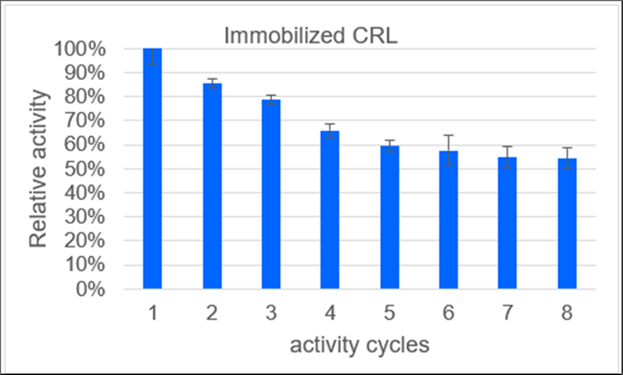
Stability assessment of free lipase and on a polypropylene membrane immobilized lipase with respect to PEM fuel cell conditions
The free and immobilized CRL (method C) were investigated further for stability with respect to storage stability and with respect to the manufacturing and operating conditions of a PEM fuel cell.
Storage stability
The examined immobilizates exhibited a residual activity of 71,6 % after 28 days of storage, as depicted in Figure 6. To elucidate the decline in enzyme activity, the supernatant solution was analyzed, revealing the presence of active enzymes. This suggests potential enzyme detachment from the membrane, notwithstanding prior washing before immobilization. Additionally, denaturation of immobilized enzymes could have occured. In contrast, free lipase enzyme activity only declined by 9.5 % over a 28 day storage period. Immobilized and free lipase show the most significant reduction of activity within the initial 14 days but remain stable afterwards. In sum, these observations underscore a general storage stability of the immobilizates for a minimum of 28 days.
Figure 6.Storage stability of the immobilizates of lipase from Candida rugosa (CRL) on polypropylene and of free CRL solution after 28 days. The highest loss of activity is observed in the first 14 days, but immobilized and free lipase stay stable in the next 14 days.
Temperature stability
As the self-healing process in PEM fuel cells is anticipated during standstill at ambient conditions, rather than operational elevated temperatures, storage experiments were conducted to assess enzyme temperature stability. Immobilized CRL samples were stored for 3 days at both 60 °C and 4 °C, maintaining a pH of 7.2, and the ensuing activity loss was quantified (Figure 7). The immobilized sample stored at 4 °C exhibited a 10.8 % activity decline, akin to the cycle-related 14.1 % loss (Figure 5). This suggests minimal activity loss during storage at 4 °C, pH 7.2. In contrast, the immobilizate stored at 60 °C for 3 days exhibited an 80.1 % loss of activity, with 14.1 % attributed to reuse (Figure 5). Notably, significant activity loss ensued, primarily due to temperature-driven enzyme denaturation. In comparison, a free CRL sample demonstrated a 95.8 % activity loss after 3 days at 60 °C. This underscores the advantageous effect of covalent immobilization on CRL, enhancing temperature stability compared to free lipase. Nevertheless, enzyme’s temperature stability is a main issue and has to be improved.
Figure 7.Activity losses of the immobilizate of lipase from Candida rugosa (CRL) on polypropylene membrane and of free CRL solution after storage for 3 d at pH 7.2 at 60 °C and 4 °C. The immobilizate is still clearly active after storage at 60°C for 3 d, despite an activity loss of 80.1 %. However, optimization is required.
Stability under acidic conditions
Given the exposure of enzymes in PEM fuel cells to pH 3 at the membrane, the impact of this acidic environment on free and immobilized lipase activity was examined (Figure 8). Immobilizates at pH 3 showed no significant difference compared to immobilizates stored at pH 7.2 (see Figure 7), yet accounting for a 3 % standard deviation. Hence, the immobilized material demonstrated stability compared to pH 3 storage, retaining activity. For activity loss determination of free CRL in an acidic solution, rebuffering to pH 7.2 via dialysis prior to p-NPA assay was essential, affecting results. During dialysis, enzyme entrapment in the membrane occurred, leading to a quantified 14.9 % loss of activity (calculation see Materials and Methods). Storing enzyme solution at pH 3 for 3 days yielded a 36.2 % substrate turnover post-rebuffering to pH 7.2. Factoring in the 14.9 % dialysis loss, the activity loss due to pH 3 treatment for free CRL was 25.0 %. Comparing free and immobilized CRL, immobilization facilitated enhanced pH stability.
Figure 8.Activity losses of the immobilizate of lipase from Candida rugosa (CRL) on polypropylene.membrane and of free CRL solution after storage for 3 d at 4°C at pH 3. The enzyme stability to acidic pH shows a significant increase by immobilizing the enzyme.
Stability against solutions containing ethanol and 1-propanol
The immobilized samples exhibit robustness against 5-minute treatments with various alcohol-containing solutions, as depicted in Figure 9. The membrane treated solely with phosphateffer (PBS) experienced a 14.4 % loss, primarily attributed to reuse of the immobilizate (14.1 %). For membranes treated with 5 % ethanol, a 21.1 % activity loss was observed, resulting in a net alcohol-induced activity decline of 7 %, after accounting for the cycle-related 14.1 % loss. In the case of the 60 % 1-propanol-treated membrane, a 37 % loss ensued due to inherent enzymes denaturation by 1-propanol. Notably, the higher concentrated 1-propanol solution markedly diminished immobilized sample activity compared to ethanol. The activity loss in the membrane treated with 5 % ethanol and 60 % 1-propanol predominantly results from 1-propanol-induced denaturation. In stark contrast, the free CRL solution displayed significantly greater activity losses, notably 91.2 % for 1-propanol. This discrepancy stems largely from the prolonged exposure of samples to alcohol in free solution, as treatment cessation is hampered by dialysis requirements.
Figure 9.Activity losses of immobilizate of lipase from Candida rugosa (CRL) on polypropylene membrane and of free CRL solution exposed to 5 % ethanol and 60 % 1-propanol containing solutions. PBS: Phosphate buffer saline. Immobilizate was treated 5 min, free enzyme solution was immidiately rebuffered after exposure, but was in contact with a deluted solution for at least 30 min. The treatment with 60 % 1-propanol caused the immobilizate to experience the highest loss of activity.
Stability against gas atmospheres
The activity of the immobilizates is not measurable affected by storage at different gas atmospheres, see Figure 10. The free CRL also shows no loss of activity due to the intercalation at different gas atmospheres. Thus, it was shown that the hydrogen and oxygen atmospheres within a PEM fuel cell do not represent a critical parameter with respect to the stability of the immobilizates.
Figure 10.Activites of lipase immobilizate from Candida rugosa (CRL) on polypropylene membran before and after storage in different gas atmospheres for 3 d and 4 °C. Values are normalized to the value of H2 measurement. The different gas atmospheres have no significant effect on the activity of the immobilizates.
Conclusion
The objective of immobilizing enzymes onto the membrane encompassed multiple facets: process establishment, analytical validation, improvement of immobilization, enhanced stability, reuse demonstration, assessment of stability under fuel cell conditions, and potential transfer to immobilization onto NafionTM.
The p-NPA assay proved effective for qualitative analysis of immobilization, especially for Candida rugosa lipase (CRL). CRL exhibited higher activity than Candida antarctica lipase B (CALB) in the assay, prompting further focus on CRL. Enzyme immobilization improvement was achieved using diisopropylcarbodiimide (DIC) added simultaneously with the enzyme. This facilitated enhanced stability under various conditions, such as temperature, reuse, and fuel cell scenarios. Demonstrably, after eight cycles, immobilized enzymes retained 54.3 % of their activity, confirming the potential for repeated self-healing. The covalent immobilization elevated CRL's temperature optimum by 10 °C.
Exposure of immobilizates to PEM fuel cell conditions demonstrated increased stability due to covalent immobilization compared to free CRL. However, activity reduction was evident and notable under specific stressors like 60 °C and concentrated alcohol solutions used in production. The immobilizates exhibited stability under pH 3 and different gas atmospheres. Considering the fuel cell's operational temperature up to 80 °C over extended periods, long-term investigations considering various conditions are crucial. Future considerations involve long-term studies simulating extended PEM fuel cell lifetimes, focusing on extremophilic enzymes or enzyme modification to enhance high-temperature stability.
The transferability to immobilization on Nafion™ will now be discussed on the basis of successful immobilization on polypropylene. While polypropylene facilitated initial investigations, Nafion™ handling complexities emerged due to its swelling behavior upon contact with aqueous media. Conventional covalent bonding was deemed unsuitable due to the proton transport function of sulfonic acid groups in Nafion™. A fluorophilic interaction approach was explored using fluorinated linker building blocks. Other results spurred development of a fluorine-labeled lysine building block for standard peptide chemistry-based enzyme immobilization on Nafion™ and further research is underway.
In conclusion, enzyme immobilization on polypropylene demonstrated enhanced stability under fuel cell operating conditions, but transfer to Nafion™ requires more specific approaches. This study is a first investigation for self-healing membranes, underscoring the need for further protein engineering to enhance temperature stability for PEM fuel cell applications. While applicable to other polymeric membranes, Nafion™ requires specific strategies due to its unique structure and function.
Acknowledgements
The authors would like to thank the Ministry of Science, Research and Arts of the Federal State of Baden-Württemberg for the financial support of the projects within the InnovationCampus Mobility of the Future (ICM). We also acknowledge the Open Access Publishing Fund of the Karlsruhe Institute of Technology.
References
- 1.Araya S, Li N, Liso V. (2022) Chapter 9 - Degradation and failure modes in proton exchange membrane fuel cells. DOI: 10.1016/B978-0-12-823708-3.00015-8. Gurbinder Kaur (Hg.): PEM Fuel Cells:.
- 2.Wallnöfer-Ogris E, Poimer F, Köll R. (2023) Main degradation mechanisms of polymer electrolyte membrane fuel cell stacks – Mechanisms, influencing factors, consequences, and mitigation strategies. , International Journal of Hydrogen Energy. DOI: 10-1016.
- 3.Jung A, Kong I, Yun C. (2017) Characteristics of hydrogen crossover through pinhole in polymer electrolyte membrane fuel cells. DOI: 10.1016/j.memsci.2016.09.009. , Journal of Membrane Science 523, 138-143.
- 4.Wang L, Advani S, Prasad A. (2016) Self-Healing Composite Membrane for Proton Electrolyte Membrane Fuel Cell Applications. , DOI: 10.1149/2.1271610jes. J. Electrochem. Soc 163(10), 1267-1271.
- 5.Böhm G, Finsterwalder F. (2004) Selbstheilende Membran für eine Brennstoffzelle. , Patent DE 103, 1.
- 6.Gartner P, Lanza G, Rudat J. (2023) Self-healing Fuel Cells by Biological Actuators. DOI: 10.1016/j.procir.202302028. Procedia CIRP 116, 161-166.
- 7.Basso A, Serban S. (2019) Industrial applications of immobilized enzymes - A review. DOI: 10.1016/j.mcat.2019.110607. Molecular Catalysis 479 110607.
- 8.Homaei A, Sariri R, Vianello F. (2013) Enzyme immobilization: an update. , DOI:, Journal of chemical biology 6(4), 10-1007.