Abstract
Co-infection of HIV with Mycobacterium tuberculosis is a common event, particularly in developing countries. The emergence and spread of multidrug resistant tuberculosis (MDR-TB) is an increasing public problem in India. The drug-resistant M. tuberculosis strains are posing a significant challenge to TB control. This study used PCR to characterize mutations inside the rifampicin resistance-determining region (RRDR) of the rpoB gene in the rifampicin-resistant M. tuberculosis co-infected with HIV. All the rifampicin-resistant strains had missense mutations. Sequence analysis detected a single or multiple hotspot mutations in the RRDR region of the rpoB gene at codons 516, 512 and 531, in most strains. Furthermore, mutations also occur at codons 512, 514, 517 and 526. The results suggest that hotspot mutations in the rpoB gene are not the sole contributors to MDR-TB co-infected with HIV.
Author Contributions
Academic Editor: Yongqiang Chen, Research associate with Dr. Spencer B. Gibson at CancerCare Manitoba, Canada.
Checked for plagiarism: Yes
Review by: Single-blind
Copyright © 2019 Shivaji K Jadhav, et al.
 This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Competing interests
The authors have declared that no competing interests exist.
Citation:
Introduction
Tuberculosis (TB) is a highly infectious disease caused by Mycobacterium tuberculosis that infects the lungs at any stage of lifecycle, and it is a major cause of death globally. HIV (human immunodeficiency virus) and TB are co-partners in crime with multidrug resistance as the major cause for the unavailability of effective treatment regimen 1. TB infection is transmitted from person to person through air by inhalation of aerosols from coughs or sprays droplets of active diseased persons. According to the World Health Organization (WHO), the death rate caused by TB is higher as compared to that caused by other diseases triggered by a single infectious agent. Worldwide, in the year of 2017, M. tuberculosis caused around 1.3 million deaths and 10 million new TB cases out of which 5.8 million was men, 3.2 million women and 1.0 million children. 2 Multidrug resistant TB (MDR-TB) results from resistance to one of the two most active first-line drugs rifampicin (RMP) and isoniazid (INH)3.
In the year of 2017, around 558,000 multidrug resistance cases were registered out of which in India (24%), China (13%) and Russian Federation (10%)2 (WHO report, 2017). This was caused by inappropriate treatment, lack of knowledge towards complete treatment; indigence and improper healthcare facilities 4. Peoples with low immune system are at the high risk. Mutations in the 81bp RRDR (rifampicin resistance determining region) of rpoB gene are the main reason for M. tuberculosis resistance to rifampicin. More than 95% mutations only occur in this 81 bp hotspot region (codons 507 to 533).5, 6 The Proper treatments with optimal time and drug dosing is a key factor to prevent drug resistance along with maximum efficacy. Rifampicin interacts with β subunit of RNA polymerase (RNAP) to arrest DNA directed RNA synthesis in M. tuberculosis. Spacer oligonucleotide typing also called as spoligotyping is a PCR based method of genotyping which is commonly used in detection of M.tuberculosis complex. This method amplifies the whole DR region (direct repeats) to check the DNA polymorphism in the spacer sequences. As the DR region is highly conserved in M. tuberculosis, it is used to differentiate different strains 7. In the case of disease outbreaks or emergency, Spoligotyping is very rapid and simple as compared to other genotyping methods 8, 9. Mutations in the rpoB gene can cause bacterial resistance to rifampicin which can use as a surrogate marker for the detection of multidrug resistance 10.
In this study, we used PCR and sequencing techniques to identify point mutations within the 81 bp hotspot RRDR in clinical samples collected from different health care centers in Hyderabad. These samples were also subjected to spoligotyping to identify the drug resistant strains.
Materials and Methods
Sample Size
We have collected 22 samples of HIV infected individuals co-infected with from primary health centre in Hyderabad and among the collected samples few were acid-fast bacilli (AFB) positive and some of them were positive by culture method. It is extremely difficult to obtain MDR cases with HIV co-infected individuals. We have screened for MDR-TB and HIV co-infection but out of 22 patients 13 were false culture positive and we excluded them from the study. Thus, a total 9 samples were screened for point mutations within the 81 bp RRDR using PCR and DNA sequencing methods. The generated sequences were comparing with wild type M. tuberculosis (H37Rv) strain to check for any mutations within the hotspot region. Finally, these samples were used for spoligotyping to identify the resistance strains.
Genomic DNA Extraction
All the sputum samples were first treated with N-acetyl-l-cysteine–sodium hydroxide (NALC-NaOH) to inactivate the pathogen 11, 16After the treatment DNA extraction was performed using QIAamp DNA Mini Kit (Qiagen) to enhance the DNA yield as per the manufacturer's instructions. Extracted DNA was measured using the Nanodrop ND-1000 Spectrophotometer (Thermofisher) and visualized on 0.8% agarose gel stained with ethidium bromide.
DNA Amplification & Sequencing
The 81bp hotspot RRDR region of rpoB gene was amplified by polymerase chain reaction (PCR) using the primers: rpoB Forward, 5’-GGATCAGCTCGCCGACCGTA-3’ and rpoB Reverse, 5’-TACGGCGTTTCGATGAACC-3’ mentioned 16. The reaction mixture contains 0.4 μM of each primer and Taq polymerase 2X master mix (G-Bioscience) ( Taq polymerase supplied with 2X Taq buffer, 0.4mM dNTPs, 3.2mM MgCl2 and 0.02% bromophenol blue). Applied Biosystem thermal cycle was used for the PCR reaction under the following conditions, initial denaturation at 95⁰C for 2 min; 34 cycles of 95⁰C for 40 sec, 55°C for 50 sec and 72⁰C for 30 sec; and a final elongation at 72⁰C for 7 min, followed by holding at 4°C. M. tuberculosis H37Rv was used as a positive control and Nuclease free water as a negative control. After amplification the samples were run on 1.5% agarose gel for visualization of amplified DNA. The amplified PCR products were purified with QIAgen PCR purification kit (Qiagen) and used as a template for cycle sequencing reactions. Both the strands of each product were sequenced using the same primers which are used for PCR amplification using ABI Big Dye Terminator v.3.0 (Applied biosystem). The cycle sequencing PCR was carried out under the following conditions: 96°C for 1 min, 96⁰C for 10 sec, 50°C for 5 sec and 60⁰C for 4 min for 30 cycles, followed by holding at 4°C. The cycle sequencing products were purified using ethanol/EDTA precipitation protocol as per the manufacturer's instructions and sequenced using ABI 3500 Genetic Analyzer.
Spoligotyping
Genomic DNA was extracted and used for amplification of spacer sequence with the labeled primers to amplify the whole DR region using the commercially available kit (Mapmygenome, Hyderabad) according to a standardized method using the designated primers of DRa and DRb 7, 12 The reaction mixture contains 0.6μM of each primer and Taq polymerase 2X Premix Taq (Takara) (in which Taqpolymerase supplied with 2X Taq buffer, 0.4mM dNTPs, 3 mM MgCl2). PCR was carried out under the following conditions: initial denaturation at 94⁰C for 3 min; 25 cycles of 94⁰C for 1 min, 55°C for 1 min and 72⁰C for 30 sec; and a final elongation at 72⁰C for 7 min, followed by holding at 4°C. Nuclease free water was used as negative control along with M. tuberculosis H37Rv and M. Bovis as positive controls, provided in the kit. After amplification the samples were run on 1.5% agarose gel for visualization of amplified DNA. The PCR products generated are biotinylated due to one of the primer biotin labeled are hybridized with the 43 immobilized spacer oligos with known sequences as per the manufacturer’s instruction. After incubation with the chemiluminescence system (GE Healthcare, UK), the hybridized fragments were visualized as a black squares on the film. The spoligotyping result was recorded in the form of binary code for positive hybridization recorded as 1 and the negative hybridization as 0. Finally, the binary codes were checked for the families & lineage using TB-VIS online tool.
Results
All the rifampicin-resistant strains had missense mutations. Sequence analysis detected a single or multiple mutations in the RRDR region of the rpoB gene at codons 516, 512 and 531. However, few MDR-TB isolates with HIV co-infection did mot harbour mutations in the RRDR hotspot region. We have observed that apart from these hotspot mutations other mutations at codons 512, 514, 517 and 526 were observed, indicating that high frequency of mutations occurs in HIV co-infected individuals. DNA extracted from 9 samples was checked on the agarose gel stained with ethidium bromide for confirmation of positive bands. All samples showed positive bands. The extracted DNA samples were further used for rpoB amplification. After amplification all products were checked on 1.5% agarose gel for 411 bp positive amplification (Figure 1).
Figure 1.Agarose gel electrophoresis 1.5% gel showing product size 411 bp. (Well no. 1-10 PCR products from sample no. 1 to 9 , except the well no.3 is blank), (well no 11- blank, 12- Negative control, 13- Positive control (M. tuberculosis H37Rv)), (M- 100 bp marker)
Sequencing of the PCR purified samples was performed by capillary electrophoresis. The results were screened for mutations in the rpoB gene with reference to M. tuberculosis H37Rv strain sequence. In drug resistance pattern, there is an amino acid substitution or amino acid change at the codon position 531 in 81 bp hotspot region of RRDR in the rpoB gene. It is known that the most common mutation in the rpoB gene is at codon 531 with serine to leucine substitution (TCG to TTG) 14. All samples were aligned with M. tuberculosis H37Rv reference sequence for mutation screening in rpoB gene out of which only one sample (Sample-9) shows the mutation at codon 531 (Figure 2). In Figure 3, the highlighted nucleotide of the Sample-9 represents the T nucleotide while that in the remaining samples represents the C nucleotide. Figure 4 shows the nucleotide sequence chromatogram. Figure 4a shows no change in the nucleotide at codon position 531 (TCG) while Figure 4b shows the nucleotide change at the codon position 531 (TCG to TTG i.e. Serine to Leucine). We have also screened for all possible codons for associated mutations (Figure 2) but we didn’t find any mutations except the Sample-9 shows serine to leucine mutation (TCG to TTG), thus indicates MDR resistance.
Figure 2.Nucleotide sequence aligned with gi|448814763:759807-763325 M tuberculosis H37Rv, reference sequence for screening of rpoB gene mutations out of which only sample-9 shows mutation at codon 531.
Figure 3.Sequence alignment using Clustal W in MEGA7 software tool.
Figure 4a.Mutation screening at codon position 531 of 81 bp (RRDR) region of rpoB gene (531 position TCG - No change indicates not a drug resistance)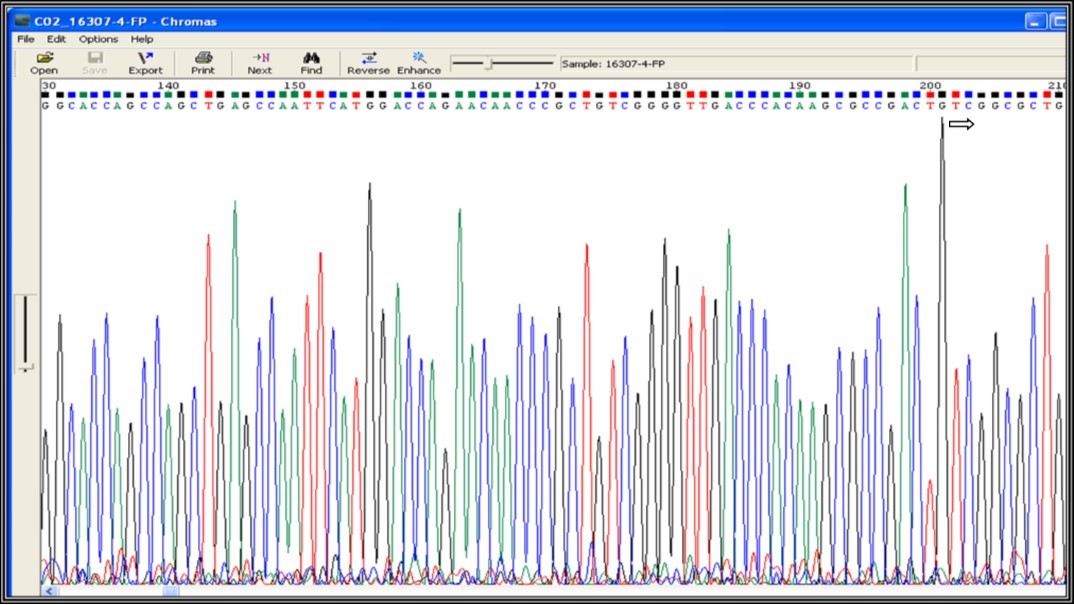
Figure 4b.Mutation screening at codon position 531 of 81 bp (RRDR) region of rpoB gene (531 position TCG – TTG i.e. Serine – Leucine change indicates drug resistance)
The same DNA samples used for sequencing were analyzed by spoligotyping. All the extracted DNA samples were used for amplification of DR region. After amplification all products were checked on 1.5% agarose gel (Figure 5). Spoligotyping of 9 samples shows distinct spoligopatterns (Figure 6). Out of the 9 samples, 4 samples (44.5%) belongs to M. tuberculosis EAI3 family, 2 samples (22.2%) belongs to M. tuberculosis CAS and the others samples belongs to M. tuberculosis EAI5 (11.1%), M. tuberculosis Beijing (11.1%), and M. bovis-BCG (11.1%). The families and lineage were identified and assigned using TB-VIS online tool. In this study, 5 samples were from Indo-Oceanic lineages (55.5%), 1 sample from M. bovis (11.1%), 1 sample from East Asian (Beijing) (11.1%) and 2 samples from unknown lineages (22.2%). The details of spoligotyping defined lineage and families of study samples were shown in Table 1 and Table 2.
Figure 5.Agarose gel electrophoresis 1.5% gel showing amplification of DR region. (M- 100 bp marker Well no. 1-9 PCR products from sample no. 1 to 9, well no 10- Positive control(M. tuberculosis H37Rv), 11- Negative control.
Figure 6.Spoligotyping hybridization pattern of amplified mycobacterial DNAs of 9 samples and 2 controls M.tuberculosis H37Rv, M.bovis BCG P3. Dark spots represent the positive hybridization & empty spots represent no hybridization i.e. absence of spacers.
| Sample IDs | Spoligotyping Binary Code | Lineage | Family |
| Sample-1 | 1101111101111110111111111111111111111100000 | Mycobacterium bovis | M. bovis-BCG |
| Sample-2 | 1001111111111111111111111111000010110001111 | Indo-Oceanic | M. tuberculosis EAI3 |
| Sample-3 | 0000000000000000000000000000000000111111111 | East Asian (Beijing) | M. tuberculosis Beijing |
| Sample-4 | 0110000111111111111111000000000000111111111 | Unknown | M. tuberculosis CAS |
| Sample-5 | 1001111111111111010001111111000010110001111 | Indo-Oceanic | M. tuberculosis EAI3 |
| Sample-6 | 1111111111111111111111111111000010111110111 | Indo-Oceanic | M. tuberculosis EAI5 |
| Sample-7 | 1110000111111111111110000000000000111111111 | Unknown | M. tuberculosis CAS |
| Sample-8 | 1001111111111111111111111111000010110001111 | Indo-Oceanic | M. tuberculosis EAI3 |
| Sample-9 | 1001111111111111111111111111000010110001111 | Indo-Oceanic | M. tuberculosis EAI3 |
| Antituberculous agents | Gene | Size (bp) | Product | Mutationfrequency among resistant MTB isolates (%) |
| Rifampicin | rpoB | 3,534 | p subunit of RNA polymerise | > 95 |
| Isoniazid | katG oxyR-ahpC inhA kasA | 2,205 585810 1,251 | Catalase-peroxidase Alkylhydroreductase Enoyl-ACP reductase 13-Ketoacyl-ACP reductase | 60-70 ~ 20 <10 <10 |
The samples used in this study were also determined for Phylogenetic analysis using Maximum Likelihood method 13 and distance matrix constructed using MEGA7software tool. In Figure 7, the numbers at the nodes are the bootstrap values based on percentage. The numbers of base substitutions per site between sequences are shown in Figure 8. The analyses were conducted using the maximum composite likelihood model for study samples.
Figure 7.Molecular Phylogenetic analysis by Maximum Likelihood method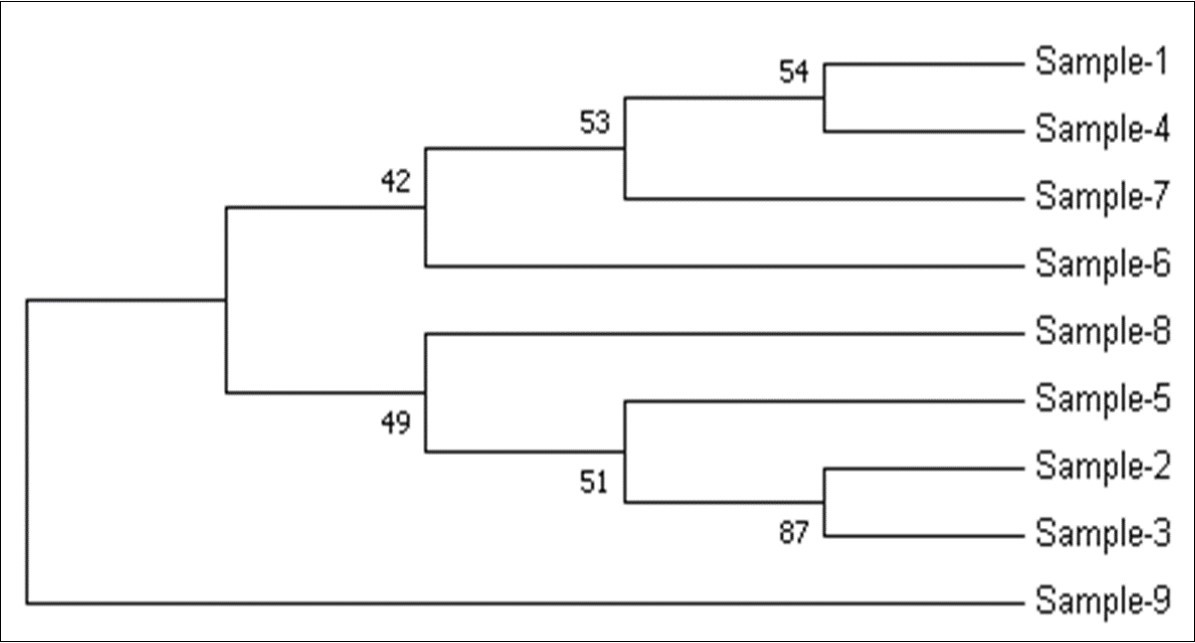
Figure 8.Estimates of evolutionary divergence between sequences
Discussion
In this study, we used DNA sequencing for the screening of MDR-TB from the collected clinical samples from different healthcare centres in Hyderabad and Spoligotyping for lineage and families’ identification. Majority of the MDR-TB isolates had mutations in the rpoB gene, and 88% of these mutations were located in three codons (531, 526 and 516) at the 81 bp hotspot region which contributes to drug resistance 15.
The rpoB gene is considered as a reliable marker accounting for more than 95% drug resistant strains of tuberculosis but other genes like katG (in more than 70 drug resistant strains) are also a suitable biomarker for MDR resistance. Screening for other genes is required for further confirmations.
As per increasing incidence of TB patients in developing countries and the emergence of MDR-TB, DNA sequencing is a very simple, cost-effective, and rapid method for the diagnosis before clinical treatment for patients. Sequencing along with Spoligotyping improves the identification of M. tuberculosis (MTB) isolates. The conventional methods like sputum microscopy and chest X-ray for detection and diagnosis for antibiotic sensitivity are time consuming as compared to molecular methods 5.
The results from this study suggest that mutations in the rpoB gene hotspot may not be the only factors associated with rifampicin resistance in TB co-infected with HIV. Mutations at other sites may also contribute to drug resistance. Therefore, GenexPert focusing on targeted hotspot regions may miss out mutations at other sites. Further studies about these novel mutations in addition to those in the hotspot region in the TB-HIV co-infected population would add more diagnostic markers for the reliable prediction of drug resistance. This will provide insight into developing novel diagnostic tools for the detection of M. tuberculosis co-infected with HIV in a high TB-HIV endemic area like India.
References
- 1.Thirumurugan R, Kathirvel M, Vallayyachari K, Surendar K, Antony S. (2015) Molecular analysis of rpoB gene mutations in rifampicin resistant Mycobacterium tuberculosis isolates by multiple allele specific polymerase chain reaction in Puducherry. , South India.J Infect Public Health 8(6), 619-625.
- 3.Sharma S, Madan M. (2014) Detection of Mutations inrpoBGene of Clinically Isolated M.tuberculosis by DNA Sequencing.JMycobacDis.(4),156.
- 4.Pandey S, Lamichhane A, Byanjankar A, Kharel A, Rai C.et al.(2017) Direct detection of rpoB and katG gene mutations of Mycobacterium tuberculosis in clinical samples.Asian. , Pac J Trop 7(8), 698-701.
- 5.Makadia S, Jain A, Patra S, Sherwal B L, Khanna A. (2012) Emerging Trend of Mutation Profile of rpoB. , Gene in MDR Tuberculosis, North India.Indian J ClinBiochem 27(4), 370-374.
- 6.Ma X, Wang H, Deng Y, Liu Z, Xu Y. (2006) rpoB Gene Mutations and Molecular Characterization of Rifampin-Resistant Mycobacterium tuberculosis isolates from Shandong Province. , China.J ClinMicrobiol 44(9), 3409-3412.
- 7.Kamerbeek J, Schouls L, Kolk A, M van Agterveld, van.Soolingen D,et al.(1997) Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology.J ClinMicrobiol. 35(4), 907-914.
- 8.Suzana S, Shanmugam S, Uma Devi KR, Swarna Latha PN. (2017) Spoligotyping of Mycobacterium tuberculosis isolates at a tertiary care hospital in India.Trop Med Int Health. 22(6), 703-707.
- 9.Gori A, Bandera A, Marchetti G, Degli Esposti A, Catozzi L. (2005) Spoligotyping and Mycobacterium tuberculosis.EmergInfect Dis. 11(8), 1242-1248.
- 10.Siddiqi N, Shamim M, Hussain S, Choudhary R K, Ahmed N. (2002) Molecular characterization of multi drug resistant isolates of Mycobacterium tuberculosis from patients from north India.AntimicrobAgents Chemother. 46(2), 443-450.
- 11.Mercy A L, Srikantam A, Jain S, KVSM Rao, Rao R. (2010) Clinical and geographical profiles of rpoB gene mutations in Mycobacterium tuberculosis isolates from Hyderabad and Koraput in India.JMicrobiolAntimicrob. 2(2), 13-18.
- 12.Chawla K, Kumar A, Shenoy V P, Chauhan D S, Sharma P. (2018) Genetic diversity of Mycobacterium tuberculosis in south coastal Karnataka, India, using spoligotyping.Indian. , J Med Res 147(3), 278-286.
- 13.Kumar S, Stecher G, Tamura K. (2016) Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets.MolBiolEvol. 33(7), 1870-1874.
- 14.Abdelaal A, El-Ghaffar H A, Zaghloul M H, N El Mashad, Badran E et al. (2009) Genotypic detection of rifampicin and isoniazid resistant Mycobacterium tuberculosis strains by DNA sequencing: a randomized trial.Ann ClinMicrobiolAntimicrob. 8(5).
