Abstract
Following ocular trauma and retinal detachment, gliotic changes in the retina may develop over the subsequent month, a process known as PVR (proliferative vitreoretinopathy). There have been no successful therapeutic interventions to inhibit PVR. The protein CTGF (Connective Tissue Growth Factor) has been associated with retinal PVR and other fibrotic diseases of the retina in clinical studies but the mechanistic link between different pathologies and retinal gliosis has not been determined. In addition, CTGF has been previously noted to be associated, in some cases, with YAP/TAZ (Yes-associated protein and Tafazzin protein complex), transcriptional regulatory proteins that change subcellular localization in response to mechanical cues, such as the stiffness of the underlying material. We have previously shown that the mRNA for CTGF is markedly (100-fold) upregulated in retinal Müller cells grown on soft substrates.
In order to evaluate if the mechanism by which mechanotransduction modulating CTGF production in retinal Müller cells involves the YAP/TAZ complex, this study tests the influence of substrate stiffness on the time dependence of CTGF protein expression, as well as subcellular localization of YAP/TAZ using a conditionally-immortalized mouse retinal Müller cell line plated on laminin-coated, polyacrylamide substrates of varying elastic modulus. Changes were assayed using immunohistochemistry and ELISA (Enzyme-Linked ImmunoSorbent Assay).
In retinal Müller cells, the relationship between elastic modulus and the pattern of CTGF protein expression was bimodal, with CTGF levels rising more rapidly for cells on hard substrates and more slowly for cells grown on soft substrates. In addition, nuclear localization of YAP/TAZ corresponded directly to the maximum CTGF expression.
Author Contributions
Academic Editor: Rajajeyakumar Manivel, Chennai Medical College Hospital & Research Centre (SRM Group), Email: [email protected]
Checked for plagiarism: Yes
Review by: Single-blind
Copyright © 2018 Joshua T Davis, et al.
 This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Competing interests
The authors have declared that no competing interests exist.
Citation:
Introduction
Extracellular matrix stiffness plays a critical role in influencing the morphology, gene expression, and differentiation of a wide variety of cell types1, 2, 3, 4, 5, 6. The elastic modulus of the cells in adult mammalian retina varies between 200 Pa and 1000 Pa, and many pathologic processes of the retina lead to local changes in stiffness of retinal tissue7. For example, Bruch’s membrane increases in stiffness by an order of magnitude with age8. Retinal detachments and laser photocoagulation can also strongly affect the stiffness of the retina16.
At the molecular level, Connective Tissue Growth Factor (CTGF) expression is directly regulated by Yes-associated protein and Tafazzin protein (YAP/TAZ) complexes9, 10. Previously, substrate stiffness was shown to alter the nuclear vs. cytoplasmic localization of the YAP/TAZ protein complex leading to the hypothesis that this complex may function to mediate mechanotransduction through a currently undefined pathway11. Expression of the protein CTGF was found to be strongly correlated with the localization of the YAP/TAZ protein complex in mesenchymal stem cells and endothelial cells11. As CTGF regulates multiple cellular processes, and is markedly upregulated in blinding conditions that involve retinal gliosis, such as proliferative vitreoretinopathy (PVR, retinal scarring)12, age-related macular degeneration13, 14, and diabetic retinopathy14, 15, from a clinical and translational standpoint, understanding the mechanical influence of expression of CTGF will be helpful in developing novel therapies for retinal disease.
Previously, we demonstrated that Müller cells increase in spread area and change levels of mRNA transcription of CTGF with increased substrate stiffness16. Based upon these observations, we used the same conditionally immortalized Müller cell line17 to determine the changes in YAP/TAZ complex localization and CTGF protein expression of as a function of time and substrate stiffness.
The goal of the present study was to understand how substrate stiffness influenced the time course of CTGF expression and YAP/TAZ localization in a conditionally immortalized Müller glial cell line17.
Materials and Methods
Cell Culture
Conditionally immortalized Müller cell (ImM10)17 were plated on laminin-coated polyacrylamide gels18,19 or glass substrates and maintained in cell culture at 5.5% CO2. After plating, adherent cells were maintained for 24 days at 39ºC in growth medium under nonimmortalizing conditions (Neurobasal media with 2% FBS, 1x B27 supplement, 2 mM L glutamine, 5 U/ml penicillin, and 5 ug/ml streptomycin (Gibco/Invitrogen)) with media changes at 3-day intervals. For growth under immortalizing conditions, media was supplemented with mouse recombinant IFN-γ (PreproTech; Rocky Hill, NJ) at 50 U/mL and cells were maintained at 33ºC. For maintenance and subsequent analyses, cells were grown on uncoated tissue culture dishes under immortalizing conditions, unless otherwise stated.
Substrate Preparation
Glass coverslips were coated with a drop of 3-aminopropyltrimethoxysilane spread evenly on the surface. Each coverslip was then washed with deionized water, placed into foil packets, and autoclaved. Once autoclaved, the coverslips were transferred to a sterile petri dish containing 0.5% gluteraldehyde (70% stock solution, Sigma) in PBS and incubated for 30 minutes at room temperature. This was followed by 5, 10 minute washes in autoclaved deionized water. After the last wash, the coverslips were laid out onto a piece of autoclaved foil in a biosafety cabinet to dry.
Gel Formation
Three different stiffnesses of gel were prepared: 5000, 1000, and 500 Pa, using mixtures of acrylamide and bisacrylamide and elastic moduli were confirmed using atomic force microscopy and rheometry. Mixtures used to generate substrates of each stiffness were as follows: 5000 Pa: 7.5% acrylamide 0.5% bisacrylamide; 1000 Pa: 7.5% acrylamide 0.2% bisacrylamide; 500 Pa: 7.5% acrylamide 0.1% bisacrylamide. Following addition of TEMED, mixtures were sterilely filtered using a 0.22 micron syringe filter. Each mixture (5.9 µL) was placed onto an individual coverslip, which was then covered with a second sterile coverslip that had been pre-treated with Sigmacote (Sigma, St Louis, MO), according to the manufacturer’s instructions, to ensure even spreading of the acrylamide. After polymerization, the overlying coverslip was removed and gels, on their base coverslips, were placed in a 24-well plate, washed with HEPES and incubated in SULFO-SANPAH (0.5 mg/ml)(Sigma). The polymer surfaces, immersed in SULFO-SANPAH, were then activated under UV light for 10 minutes, followed by three more washes in HEPES buffer. Immediately after removal of the wash, 200 µL of laminin (10 µg/ml in HEPES) was added to each well to cover the acrylamide substrate and incubated at 37ºC (5.5% CO2) for 4 hours. Subsequently, all liquid was removed and replaced with growth media and plates were returned to incubate at 37ºC for at least 15 minutes before cells were added.
Immunohistochemistry
For immunostaining, cells were fixed in 4% paraformaldehyde for 10 minutes at 37ºC. Samples were washed in PBS, incubated in PBS containing 0.5% Triton X-100 (10 minutes), and non-specific binding was blocked by incubating the samples in blocking buffer (PBS with 10% normal goat serum/0.5% Triton X-100/1% fish gelatin/5% bovine serum albumin) for 1 hour. Primary antibodies to YAP/TAZ (4912S, Cell Signaling Technology) were diluted 1:200 in blocking buffer and applied for 2 hours at room temperature on a rotary platform. Secondary antibodies, conjugated to Alexa Fluor 555 (A21429, Gibco/Invitrogen) were diluted 1:500 and incubated with the samples for 2 hours at room temperature. Samples were then mounted using Slowfade Gold with DAPI (S36938, Invitrogen). Specificity of labeling was confirmed by omitting primary antibody or by substituting normal serum for the species used to generate the primary antibody. Immunostained cells were imaged with an inverted microscope (IX71; Olympus, Tokyo, Japan) with monochrome, cooled CCD digital camera (Rolera-XR; Q-Imaging, Surrey, BC, Canada). Three biological replicates were performed. All images were photographed at the same exposure time.
ELISA
ELISA was performed as a sandwich enzyme immunoassay using ELISA plates prepared with antibodies against mouse CTGF (SEA010Mu, USCN Life Sciences, Inc, Wuhan, China) and according to the manufacturer’s instructions. ImM10 cells were cultured in 35mm dishes on the four different substrates for 24 days, with complete removal of the supernatant every 3 days for the first 12 days and then every 4 days until day 24. Each supernatant sample was transferred to a microcentrifuge tube and frozen at -80ºC until all samples were collected. All samples were collected from three independent experiments and diluted 1:20 in deionized water before use in the ELISA assay. Prior to being transferred to the substrates of different elastic modulus, the cells were initially cultured, under non-immortalizing conditions, on 1000 Pa gels at 37ºC for 14 days.
Results
Müller cells cultured on the different substrates demonstrated two distinct temporal patterns of CTGF protein expression (Figure 1). On firmer substrates (glass and 5000 Pa gels), the maximal protein expression occurred at day 16 in culture, whereas on softer substrates (1000 Pa and 500 Pa), the maximal protein expression occurred at day 20. Although there were no statistically significant differences within groups (glass vs. 5000 Pa or 1000 Pa vs. 500 Pa), differences between the firmer and softer substrates were significant (2-way ANOVA, p <0.0001). To further analyze these findings, and the relationship between CTGF levels and subcellular localization of the YAP/TAZ complex, immunohistochemistry for YAP/TAZ was performed on cells cultured for 16 (Figure 2) or 20 days (Figure 3) on 500 Pa, 1000 Pa, 5000 Pa, and laminin-coated glass substrates. The proteins YAP and TAZ were found to be predominantly nuclear only at the time of maximal CTGF protein expression.
Figure 1.Graph showing ELISA quantification of CTGF protein as a function of both time and substrate stiffness. CTGF expression reaches a maximum earlier (16 days in culture) on firmer substrates relative to 20 days for the softer substrates. Each point shows the mean of 3 biological replicates. Error bars denote the SEM. This finding is significant (two-way ANOVA: p <0.0001).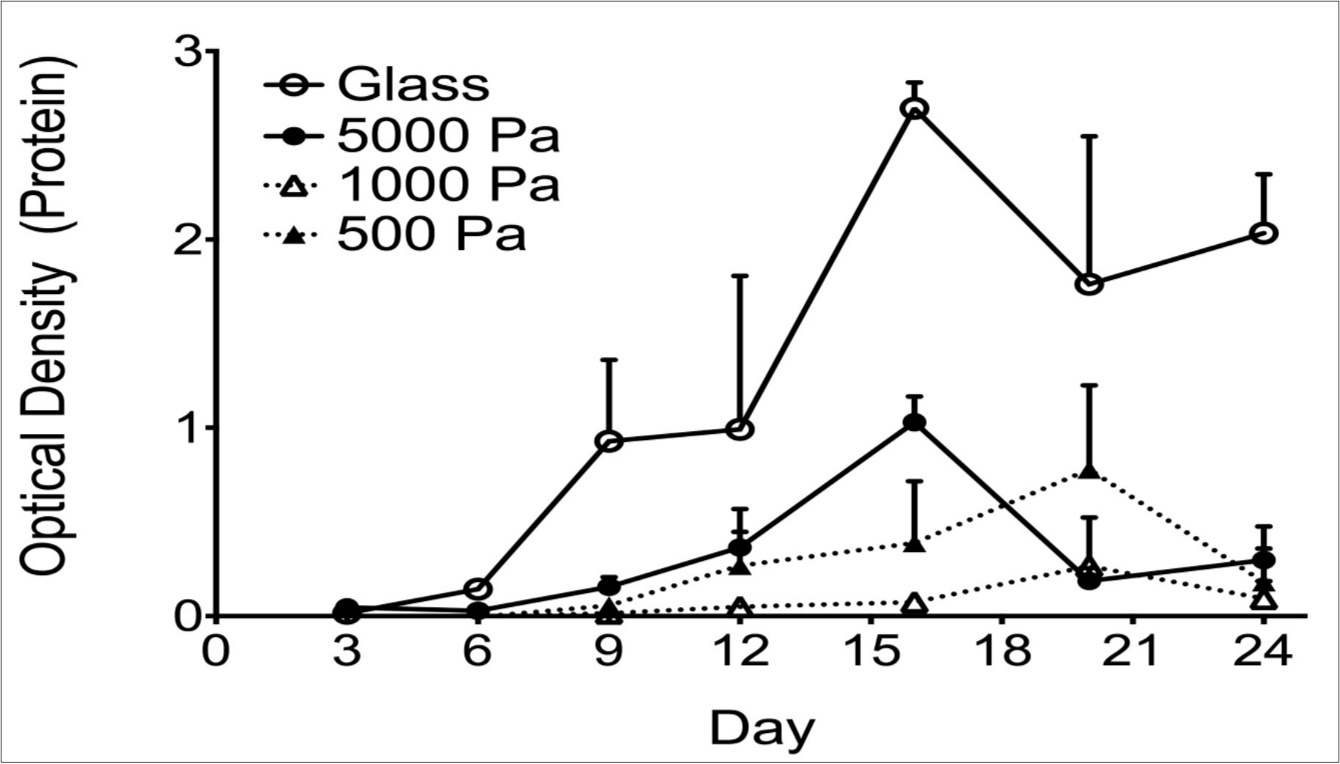
Figure 2.(A-F) Immunohistochemistry for the YAP/TAZ complex in Müller cells at 16 days in culture. The YAP/TAZ complex is localized to the cytoplasm of cells grown on 500 Pa (A, B) and 1000 Pa (C, D) gels, as demonstrated by the corresponding DAPI staining of DNA (A’, B’, C’, D’). The YAP/TAZ complex in cells grown on 5000 Pa gels (E, F) is localized to the nucleus, as shown by DAPI stain (E’, F’). Scale bar: 50 microns.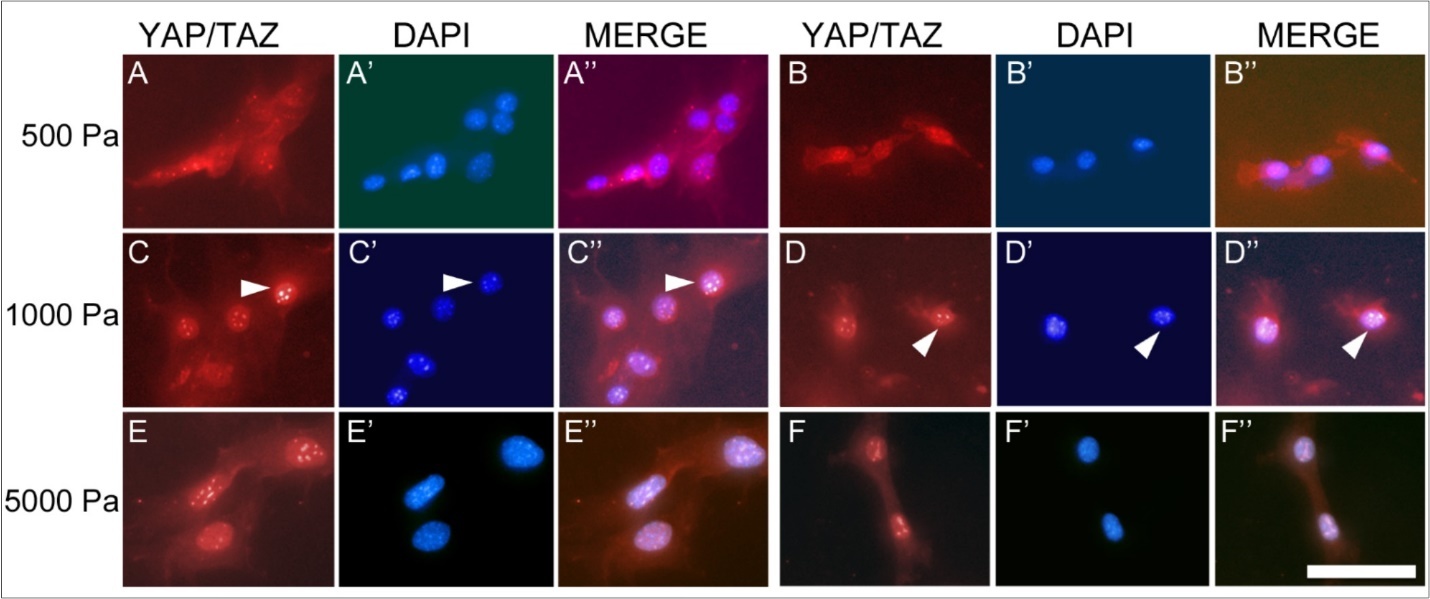
Figure 3.Immunohistochemistry for the YAP/TAZ complex in Müller cells at 20 days in culture. The YAP/TAZ complex is localized to the nucleus in cells grown on 500 Pa (A,B) and 1000 Pa (C, D) gels as shown by the corresponding DAPI staining of nuclear DNA (A’, B’, C’, D’). The YAP/TAZ complex in cells grown on 5000 Pa gels (E, F) is primarily localized in the cytoplasm as demonstrated in the corresponding DAPI stain (E’, F’). Scale bar: 50 microns.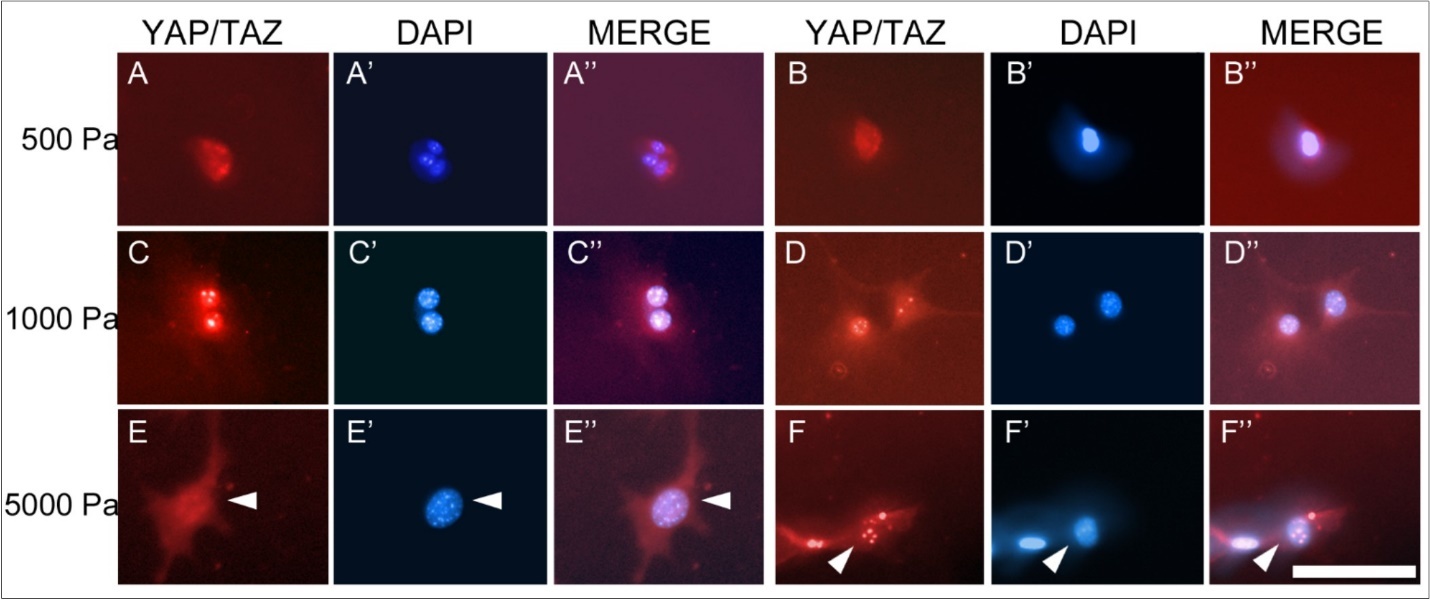
Discussion
There is growing evidence11 that cellular response to substrate stiffness is a bimodal function of stiffness, and that this bimodal response is at least in part a result of changes in the subcellular trafficking of YAP/TAZ complexes between the cytoplasm and nucleus. Previously, we noted that cortical actin stiffness, as measured by AFM, increased linearly up to a substrate stiffness of 2000 Pa, and then did not change further16. In addition, Müller cells on substrates of 1000 Pa or less proliferated at a slower rate than cells on firmer substrates16. Given that the normal, physiological stiffness of the retinal neurons that surround Müller cells is ~1000 Pa7, these data suggest that the transition point between ‘hard’ and ‘soft’ for this cell type corresponds well to their natural environment. A small change in tissue stiffness away from equilibrium has the potential to induce substantial changes in cell behavior. This finding also suggests that CTGF and YAP/TAZ may play a role in tissue stiffness homeostasis through a feedback loop in which tissue that becomes overly soft will produce more CTGF, which will act to stiffen the tissue16.
Our finding that the time-dependence of YAP/TAZ localization is influenced by substrate stiffness can account for the difference in time of maximal expression of CTGF in cells grown on substrates of different stiffness. Experimentally, our results suggest that authors who publish levels of gene expression of cells on different substrates should consider the time that the cells have been in culture.
If we compare other similar studies (see Table 1), we notice a trend in that ‘soft’ substrates are similar in stiffness to the cell’s normal, physiologic environment, and that the physiological environment of different cells may differ by more than an order of magnitude. Together, these data suggest that each cell type responds similarly across a range of physiologically ‘normal’ elastic moduli, above which its environment is considered ‘stiff’ and the cellular response changes. This is consistent with observations from this and other studies11, 20 showing that cellular responses to changes in environmental stiffness tend to be bimodal rather than continuous. The dramatic differences in physiological stiffness between different tissues and the range of responses of different resident cell populations suggest that cells have intrinsic detection mechanisms and biomechanical responses that are likely determined during tissue development and maturation.
Table 1. A review of similar mechanotransduction studies in which the investigators have determined a ‘soft’ and ‘firm’ regime experimentally. Note that the ‘soft’ regime is typically close to the normal, physiologic stiffness of the tissue that the cells encounter in vivo as well as the range of what cells consider a normal stiffness.| Cell Type | Physiologic | Soft | Firm |
| Astrocytes | 0.33 | <1 | >2 |
| Hepatic stellate cells | 0.3-0.6 | 0.4-11 | 1.75-12 |
| Müller Cells16, 7 | 1 | <2 | 5 |
| Trabecular Meshwork Cells | 5 | 5 | 75 |
| Corneal fibroblasts, | 25 | 10 | 85 |
Although CTGF is one gene that is strongly regulated by the YAP/TAZ11, Ankyrin repeat domain-containing protein 1 (ANKRD1), a transcription factor that may be involved in the myofibrillar stretch-sensor system, is also strongly regulated by YAP/TAZ11, and future clinical studies might evaluate the role of this protein in retinal disease.
Conclusion
In summary, our findings suggest that changes in local elastic modulus can have significant, time-dependent influence upon gene expression. In particular, expression of CTGF, a protein that has been previously implicated in gliotic diseases of the retina, is strongly influenced by substrate stiffness. Consideration of the local elastic modulus may thus be critical in the design and long-term integration of retinal prosthetics and stem-cell based therapies. In particular, our results may help to explain the time course for the development of PVR/retinal gliosis in retinal detachment and suggest the duration over which therapies to prevent PVR should be administered after repair of a retinal detachment. Finally, prosthetics or matrices encapsulating tissues derived from stem-cells that possess non-physiologic stiffness for the local environment in which they are placed (typically the retina and/or RPE) may induce local cellular changes to their environment that may degrade their therapeutic effect.
Acknowledgment
Research supported by the NIH/NEI (EY017112, WJF; P30 EY07551 CORE Vision Research Grant, College of Optometry, University of Houston) and a National Academies Keck Futures Grant in Advanced Prosthetics to WJF. No funds have been received to cover the cost to publish.
Abbreviations
PVR = Proliferative Vitreoretinopathy
CTGF = Connective Tissue Growth Factor
YAP/TAZ = Yes-associated protein /transcriptional coactivator with PDZ-binding motif
ELISA = Enzyme Linked Immunosorbent Assay
Pa = Pascals
FBS = Fetal Bovine Serum
PBS = Phosphate-Buffered Saline
ANOVA = Analysis of Variance
AFM = Atomic Force Microscopy
References
- 1.P C Georges, P A Janmey. (2005) Cell type-specific response to growth on soft materials. , J Appl Physiol 98, 1547-1553.
- 2.P A Janmey, J P Winer, M E, Wen Q. (2009) The hard life of soft cells. Cell Motil Cytoskeleton. 66, 597-605.
- 3.Engler A J, Sen S, Sweeney H L, Discher D E. (2006) Matrix elasticity directs stem cell lineage specification. , Cell 126, 677-689.
- 4.SFB Mennens, Bolomini-Vittori M, Weiden J, Joosten B, Cambi A et al.Substrate stiffness influences phenotype and function of human antigen-presenting dendritic cells. , Nature Publishing Group 7(1), 17511.
- 5.Mitchell M J, Jain R K, Langer R.Engineering and physical sciences in oncology: challenges and opportunities. , Nature Reviews 17(11), 659-675.
- 6.Chen Y, Ju L, Rushdi M, Ge C.Zhu C.Receptor-mediated cell mechanosensing. Weaver VM, ed , Mol Biol Cell 28(23), 3134-3155.
- 7.Lu Y B, Franze K, Seifert G, Steinhäuser C, Kirchhoff F et al. (2006) Viscoelastic properties of individual glial cells and neurons in the CNS. Proc Natl Acad Sci USA 103 17759-17764.
- 9.Zhang H, C Y Liu, Z Y. (2009) TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. , Journal of Biological Chemistry 284(20), 13355-13362.
- 10.K C Lin, Moroishi T, Meng Z. (2017) Regulation of Hippo pathway transcription factor TEAD by p38 MAPK-induced cytoplasmic translocation. , Nature Cell Biology 19(8), 996-1002.
- 11.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S et al. (2011) Role of YAP/TAZ in mechanotransduction. Nature. 474-179.
- 12.D R Hinton, He S, M L Jin, Barron E, S J Ryan. (2002) Novel growth factors involved in the pathogenesis of proliferative vitreoretinopathy Eye. 16, 422-428.
- 13.Nagai N, Klimava A, W H Lee, Izumi-Nagai K, J T Handa. (2009) CTGF is increased in basal deposits and regulates matrix production through the erk (p42/p44mapk) mapk and the p38 signaling pathways. Invest Ophthalmol Vis Sci. 50, 1903-1910.
- 14.He S, Chen Y, Khankan R, Barron E, Burton R et al. (2008) Connective tissue growth factor as a mediator of intraocular fibrosis. Invest Ophthalmol Vis Sci. 49, 4078-4088.
- 15.E J Kuiper, R van Zijderveld, Roestenberg P, K M Lyons, Goldschmeding R et al. (2008) Connective tissue growth factor is necessary for retinal capillary basal lamina thickening in diabetic mice. , J Histochem Cytochem 56, 785-792.
- 16.J T Davis, Wen Q, P A Janmey, D C Otteson, W J Foster. (2012) Muller cell expression of genes implicated in proliferative vitreoretinopathy is influenced by substrate elastic modulus. Invest Ophthalmol Vis Sci. 53, 3014-3019.
- 17.D C Otteson, M J Phillips. (2010) A conditional immortalized mouse muller glial cell line expressing glial and retinal stem cell genes. Invest Ophthalmol Vis Sci. 51, 5991-6000.
- 18.R J Pelham, Wang Y. (1997) Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci USA 94, 13661-13665.
- 19.Y L Wang, R J Pelham. (1998) Preparation of a flexible, porous polyacrylamide substrate for mechanical studies of cultured cells. , Meth. Enzymol 298, 489-496.
- 20.Swift J, I L, Buxboim A, Harada Takamasa, Dingal P C D P et al. (2013) Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 341(6149), 1240104-1240104.
- 21.A T Mikhailov, Torrado M. (2008) The enigmatic role of the ankyrin repeat domain 1 gene in heart development and disease. , Int. J. Dev. Biol 52, 811-21.
- 22.P C Georges, W J Miller, D F Meaney, E S Sawyer, P A Janmey. (2006) Matrices with compliance comparable to that of brain tissue select neuronal over glial growth in mixed cortical cultures. , Biophys J 90, 3012-3018.
- 23.A L Olsen, S A Bloomer, E P Chan, Gaca M D A, P C Georges et al. (2011) Hepatic stellate cells require a stiff environment for myofibroblastic differentiation. , Am J Physiol Gastrointest Liver Physiol 301, 110-118.
- 24.V K Raghunathan, T J, Dreier B, C M Reilly, S M Thomasy et al. (2013) . Role of Substratum Stiffness in Modulating Genes Associated with Extracellular Matrix and Mechanotransducers YAP and TAZ. Invest Ophthalmol Vis Sci 54, 378-386.
Cited by (7)
- 1.Zhu Xinglong, Mao Shengqiang, Yang Ying, Liu Xinmei, Liu Qin, et al, 2025, Biomimetic Topological Micropattern Arrays Regulate the Heterogeneity of Cellular Fates in Lung Fibroblasts between Fibrosis and Invasion, ACS Nano, 19(1), 580, 10.1021/acsnano.4c11113
- 2.Du Yuxiang, 2024, The Hippo signalling pathway and its impact on eye diseases, Journal of Cellular and Molecular Medicine, 28(8), 10.1111/jcmm.18300
- 3.Miller Charles G., Henderson Matthew, Mantopoulos Dimosthenis, Leskov Ilya, Greco Todd, et al, 2021, The Proteome of Preretinal Tissue in Proliferative Vitreoretinopathy, Ophthalmic Surgery, Lasers and Imaging Retina, 52(S1), 10.3928/23258160-20210518-02
- 4.Lin Lin, Yuan Yinfeng, Huang Zhihui, Wang Yongjie, 2025, YAP Signaling in Glia: Pivotal Roles in Neurological Development, Regeneration and Diseases, Neuroscience Bulletin, 41(3), 501, 10.1007/s12264-024-01308-w
- 5.Zhao Yaqin, Sun Bin, Fu Xuefei, Zuo Zhuan, Qin Huan, et al, 2024, YAP in development and disease: Navigating the regulatory landscape from retina to brain, Biomedicine & Pharmacotherapy, 175(), 116703, 10.1016/j.biopha.2024.116703
- 6.Blokland Kaj E. C., Nizamoglu Mehmet, Habibie Habibie, Borghuis Theo, Schuliga Michael, et al, 2022, Substrate stiffness engineered to replicate disease conditions influence senescence and fibrotic responses in primary lung fibroblasts, Frontiers in Pharmacology, 13(), 10.3389/fphar.2022.989169
- 7.Prieto-López Laura, Pereiro Xandra, Vecino Elena, 2024, The mechanics of the retina: Müller glia role on retinal extracellular matrix and modelling, Frontiers in Medicine, 11(), 10.3389/fmed.2024.1393057