Correlating 13C Isotope in Oligomeric Proanthocyanidins with their Anticancer Properties
Abstract
Upon considering the anticancer effects of larger oligomeric proanthocyanidins and observing various papers reporting the high resolution mass spectroscopy of the oligomeric proanthocyanidins, it is determined that the unusual 13C enrichment in some plant oligomeric proanthocyanidins may be responsible for the anticancer activities of these food products. Such correlation of the 13C in the oligomeric proanthocyanidins also correlate with their scavenging of free-radicals, anti-virial and anti-bacterial properties. Proanthocyanidins in grape seeds are observed to have high enrichment in heavy isotopes of 2H, 13C, 15N and/or 17O. Mass analysis of DNA from human cancer cells are compared to normal human cells and cancer cells show bond specific enrichment of heavy isotopes in nucleotides G, A, T and C. On such basis, this study suggests possible stronger interactions of proanthocyanidins with DNA in cancer verses DNA in normal cells due to heavy isotope bond specific enrichments in both proanthocyanidins and the cancer DNA. Such 13C interactions from oligomeric proanthocyanidins with nucleic acids and proteins involved in replications, transcriptions and translations in cancer cells for interacting and chemically altering anabolism and cell division of the cancer cells are consistent with the author’s mechanism for normal cell to cancer cell transformations via possible replacements of primordial 1H, 12C, 14N, 16O, and 24Mg isotopes by nonprimordial 2H, 13C, 15N, and 17O and 25Mg isotopes in the proteins and nucleic acids. Such is also consistent with the proposed treatment for cancer by the author by use of foods containing proteins, nucleic acids, carbohydrates and/or drug molecules enriched with the nonprimordial isotopes of 2H, 13C, 15N, and 17O and 25Mg.
Author Contributions
Copyright © 2022 Reginald B. Little et al
 This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Competing interests
The authors have declared that no competing interests exist.
Citation:
Introduction
Cancer and Metabolism
Cancer is abnormal cell reproduction exhibiting unusual metabolic processes. Cancer occurs as cells alter various normal catabolic and anabolic metabolisms. Warburg Effect involves accelerated glycolysis and suppressed Kreb cycle (catabolism). Glycolysis is catabolic process of enzymatic conversion of glucose to pyruvate. The cellular transformations to cancer lead to accelerated glycolysis. Kreb cycle is catabolic enzymatic conversion of pyruvate to carbon dioxide. The cellular transformations to cancer lead to suppression of Kreb cycle. The anabolism of genetic code is also altered during cancer formation as DNA replications and RNA transcriptions are altered (chaotically and anabolically). Such anabolic chaos with also altered consequent protein translations leads to cancer cell genesis and multiplying genetically altered cells rapidly. In this theory, the anabolic alterations of genes cause altered protein translations for producing proteins of glycolysis that accelerate glycolysis while producing proteins, associated with Kreb cycle that suppress the Kreb cycle. A big mystery of cancer is the nature and mechanism of the DNA mutation, RNA mutation and altered protein translations. In this work, the prior theory 1, 2, 3 that nonprimordial isotopes drive, DNA, RNA and protein alterations for cancer is substantiated and the use of nonprimordials to alter cancer metabolism for new treatments of cancer is further stressed.
Isotopic DNA, RNA and Protein Alterations for Mechanism
In this work, the theory 1, 2, 3 of stable isotopic replacements and substitutions of primordial, stable 1H, 12C, 14N, 16O, and 32S by nonprimordial, stable 2H, 13C, 15N, 17O, 25Mg, and 33S is further developed. This work focuses more on DNA, nucleotides and telomeres. Prior papers focused on glycolysis and Kreb cycle. In normal cells, the ends of DNA have unbounded, telomeric regions, which are shortened to terminate replications of genes, but in cancer the telomeres do not shorten and induce apoptosis. But in cancer, the telomeres mutate and involve telomerase with acceleration of replications 4. Telomerase is a protein that is associated with elongations of telomeres. It is unknown why shorten telomeres in cancer cells continue to replicate by telomerases and accelerate replications and transcriptions of DNA and RNA. In this work, the epigenetic stable-isotopic alterations by nonprimordial isotopes (2H, 13C, 15N, 17O, 25Mg, and 33S) of DNA, RNA and consequent proteins during normal cells to cancer cells transformations are proposed for fundamental chemistry of cancer’s origins and habitats 1, 2, 3 and possibly explain why the shorten telomere in cancer continue to replicate rather than terminate cell life as the shorten telomeres in normal cells.
This theory 1, 2, 3 determines that isotopic replacements in normal cells with epigenetic modifications prevent the shortening of the telomeres for causing apoptosis for causing cancer. The nonprimordial isotopes cause such alternations by interfering with signaling to apoptosis by the nonprimordials binding of the telomeres for causing consequent continued replication of the DNA with more and more replications; such that the DNA becomes too bond specifically enriched in nonprimordial isotopes (2H, 13C, 15N, 17O, 25Mg, and 33S) of different nuclear magnetic moments (NMMs) 1, 2, 3 for normal cellular functioning. But with aging of the host (unusual diet and/or external magnetism), this theory 1, 2, 3 proposes more and more biomolecules bond-specifically enrich in the nonprimordial isotopes (2H, 13C, 15N, 17O, 25Mg, and 33S) in specific bonds relative to the primordial isotopes (1H, 12C, 14N, 16O, 24Mg and 32S) for greater probability of simultaneous, multiple nonprimordial clumpings in specific bonds in both proteins and nucleic acids. On such basis, the simultaneous nonprimordials in the proteins and the DNA and RNA prevent the normal telomeric (and other gene expressions) induced cell apoptosis by primordial isotopic interactions with the proteins. The nonprimordial isotopes interacting between the telomere and telomerase prevent apoptosis for causing continued cancerous DNA, RNA, and protein reproductions and malfunctions of the normal cells to transform them to carcinomic cells by the prior theory 1, 2, 3. The prior theory 1, 2, 3 proposes that the clumpings of nonprimordial isotopes in specific bonds in the telomeres change the binding of the base pairs in the genes, so that the shorter telomeres (and indeed for other genes and their expressions) do not express apoptosis as the telomeres are bound more tightly by the nonprimordial isotopes. The telomeric genes are bound more strongly to binding proteins for telomerase expression. So that the stronger bound nonprimordial, isotopic, shorter telomeres continue to allow the DNA to replicate and the resulting nonprimordial DNA to replicate further to transcribe nonprimordial RNA and the resulting nonprimordial RNA continues to produce nonprimordial proteins. In the DNA and RNA, the accumulations of nonprimordials by 2D, 13C1H3, 15N1H2 and 17O1H (and 13C2D1H2, 15N2D21H, 17O2D) functional replacements on nucleotides of guanosine (G), adenosine (A), cytidine (C), uridine (U) and thymidine (T) rather than primordial 1H, 12C1H3, 14N1H2, 16O1H replacements cause altered, stronger bonding of the AT and GC in nonprimordial DNA and stronger, altered bonding of AU and GC in nonprimordial RNA. By the author’s model 1, 2, 3, the nonprimordial isotopes in the 2D, 13C1H3, 15N1H2 and 17O1H (and 13C2D1H2, 15N2D21H, 17O2D) on guanosine, adenosine, cytidine, uridine and thymidine cause magnetic bondings in addition to the hydrogen bondings to reduce and hinder the separations of the DNA base pairs for causing normal cells to transform to cancer cells. But by the prior theory 1, 2, 3, such can cause greater nonprimordial uptakes by the cancer DNA; so new treatments are possible as here with proanthocyanidins as the overall nonprimordial bond-specific enriched cancer DNA becomes less separable with killing of the cancer cells by over isotopically enriching the nucleic acids and proteins in the cancer.
Theory for Mechanism of Cancer and Cure 123
Atomic and Molecular Dynamics for Altered Biochemistry
The altered enzymatics of proteins and nucleic acids as by this prior theory 1, 2, 3 of cancer are based upon the different nuclear magnetic moments (NMMs) and masses of nonprimordial isotopes (2H, 13C, 15N, 17O, 25Mg, and 33S) relative to primordial isotopes (1H, 12C, 14N, 16O, 24Mg and 32S) as well as their tiny relative mass differences. Hydrogen has 2 important stable isotopes with different NMMs, spins, masses and relative abundances: 1H {99.988%, 1 ½ (I) spin, 2.79 (µ/µN) NMM} and 2D {0.0115%, 0 (I) spin, (µ/µN) ) NMM}. Carbon has 2 important stable isotopes with different NMMs, relative abundances, masses and spins: 12C {98.9%, O (I) spin, 0 (µ/µN) NMM} and 13C {1.1%, ½ (I) spin, 0.70 (µ/µN) NMM}. Nitrogen has 2 important stable isotopes with different NMMs, relative abundances, masses and spins: 14N {99.6%, 1 (I) spin, 0.40 (µ/µN) NMM} and 15N {0.4%, ½ (I) spin, -0.28 (µ/µN) NMM}. Oxygen has 3 important isotopes with different NMMs, spins, masses and relative abundances: 16O {99.8%, 0 (I) spin, 0 (µ/µN) NMM}, 17O {0.03%, 5/2 (I) spin, -1.89 (µ/µN) NMM, 18O {0.205%, 0 (I) spin, 0 (µ/µN) NMM}. Magnesium has 3 important isotopes with different NMMs, spins, masses and relative abundances: 24Mg {79.0%, 0 (I) spin, 0 (µ/µN) NMM} ,25Mg {10.0%, 3/2 (I) spin, -0.86 (µ/µN) NMM}, 26Mg {11.0%, 0 (I) spin, 0 (µ/µN) NMM}. Phosphorus has 1 important isotope: 31P {100%, ½ (I) spin, 1.13 (µ/µN) NMM}. Sulfur has 3 important isotopes with different NMMs, spins, masses and relative abundances: 32S {94.9%, 0(I) spin, 0 (µ/µN) NMM}, 33S {0.8%, 3/2 (I) spin, 0.64 (µ/µN) NMM}, 34S {4.3%, 0 (I) spin, 0 (µ/µN) NMM}.
Changes in Isotopic Abundances
This theory 1, 2, 3 proposes that the relative abundances of the unusual, uncommon nonprimordial isotopes have changed in food supplies of plants, animals and humans such that humans have increased levels of the nonprimordial stable isotopes (2H, 13C, 15N, 17O, 25Mg, and 33S) in their cells during the last 150 years for increased prevalence of cancer. The technologies of the industrial revolution, nuclear reaction uses and industry, agricultural changes, automobile technology and radio-technology are proposed by this theory 1, 2, 3 to increase nonprimordial isotopes and even redistribute isotopes into key chemical bonds in biomolecules. By the author’s theory 1, 2, 3 for instance, radiowaves are able by broad band excitations to stimulate the continua states by the author’s theory 1, 2, 3 to redistribute nonprimordial isotopes into specific chemical bonds even in normal relative abundances relative to distributions in the absence of radiowaves. Thereby with increase enrichments, the radiowaves compound the clumping of non-primordial isotopes into specific chemical bonds in proteins, RNA and DNA. Technologies introduced all these new ingredients and conditions of nonprimordials, RF and microwaves and static magnetic fields (Bext) to explain origin of cancer and acceleration of cancer.
Changes in Biomolecular Chemical Dynamics
These non-primordial isotopes reversibly, fractionally fiss and fuse to momentarily transmute to different quantum fields about the nuclei in atoms and molecules relative to the reversible, fractional fissing and fusing of primordial isotopes. Moreover, on the basis of this theory 1, 2, 3, the author has determined that the fractional, reversible fissing and fusing of the nonprimordial isotopes are more sensitive than nuclei of zero NMMs to tiny intensity surrounding fields of thermal space as by Little’s Rules 1, 2 and 3. Such reversible, fractional fissing and fusing of the stable isotopes by the author’s theory 1, 2, 3 alters the enzymatic dynamics along the reaction coordinates of protein, nucleic acid, lipid, and carbohydrate biochemical dynamics. The fractional, reversible fissing and fusing of nuclei release NMMs to surrounding electrons for ‘internal nuclear pressures’ to alter surrounding atomic orbitals and such altered atomic orbitals alter molecular orbitals and alter chemical dynamics, catalysis and enzymatics by the Little Effect: “spins alter orbitals during chemical reactions and orbitals altering spins”. The Little Effect not only involves e- spins altering orbitals but nuclear spins and nucleon orbitals also alter electronic orbitals for relativistic nuclear Little Effect as manifested by these nonzero NMMs of nonprimordials relative to more null NMMs of primordials.
For instance, the fractional, reversible fissing and fusing of the nonprimordial isotopes in enzymes can alter the stereochemistry of the substrate as the enzyme catalyzes the chemical transformation of the substrate. For instance, 14N and 15N nuclear motions have different chiralities as 14N has positive NMM and 15N has negative NMM; so changing 14N to 15N by this prior theory (1-3) would cause the fractional fissed field of 15N (relative to native 14N in the enzyme) to alter the chirality of wavefunctions from the enzymatic catalyzing transition state of the substrate relative to such fissed fields from primordial 14N. As the biomolecules have specific stereochemistry and manifest chiral environment in healthy organisms, the altered chirality can be a basis of disease as caused by 2H, 13C, 15N, 17O, 25Mg. These alterations by the author’s theory 1, 2, 3 transform normal cells to cancer cells. Such altered chemical dynamics by isotopic replacements in DNA, RNA and proteins are manifested by the accelerations of cellular reproduction, replication, transcription and protein translation with consequent acceleration of the glycolysis process and the suppression of the Kreb cycle.
On the basis of the author’s theory 1, 2, 3 the surrounding radiowaves and static magnetic fields of technologies accelerate such faster glycolysis and slower Kreb cycle. Alterations of DNA reproductions, RNA transcriptions and protein translations cause cancer. The author further notes that both technologies for more nonprimordials and technologies for more external RF, EM radiations and static magnetic fields in combination cause more cancer and continued acceleration of cancer during last 150 years. But the author notes here just as nonprimodials can affect biomolecules in normal cells to cause cancer these nonprimordials can also affect cancer itself to kill cancer and with Bext and RF such use of nonprimordials to heat and kill cancer cells is enhanced. The author in this paper uses proanthocyanidins as in skin of fruit and grape seeds as natural sources of 13C, 15N and 17O enriched polyphenols (nucleotides-like structures) to more strongly interact with cancer DNA in deadly ways (relative to normal DNA). Proanthocyanidins (PACs) are polyphenol compounds. Some foods are rich in PACs: blueberries, grapes, cranberries, cinnamon bark, hazelnuts and chocolate. Polyphenols are organic molecules having many phenol units. In this work the authors, determine similarity of polyphenol structures nucleoside structures in G, C, A, U and T nucleotides can cause favorable interactions and binding between the polyphenols of PACs and nucleotides in DNA. It is thought that the intrinsic nonprimordial bond-specific enrichment in PACs and the bond specific enrichment of nonprimordials in cancer DNA may cause stronger interactions of PACs with cancer DNA to alter cancer genetics and metabolism for use of PACs for treating cancer. The author notes the cancer already has 13C, 15N, 17O in its DNA and RNA and proteins. The proanthocyanidins have structures and 13C, 15N and 17O enriched in specific bonds to bind the causer DNA/RNA in stronger ways.
Hypothesis
In this paper, it is hypothesized that during replications and transcriptions, the primordial isotopes code active genes, but nonprimordial isotopes accumulate in inactive regions of genes. It is further hypothesized that the shorten telomeres occur in normal cells due to the accumulations of primordial isotopes in the growing telomeres and telomerases; so the primordial telomerases cannot bind as well with the shorter primordial telomeres to prevent their opening and unraveling of the telomere at ends having primordial isotopes; so in normal cells the shortened telomeres unravel at the end by the primordial isotopes to induce apoptosis. It is also hypothesized that as nonprimordial isotopes accumulate in normal cells, and DNA, RNA and proteins (like telomerase) through processes of deuterations, methylations, aminations, hydroxylations and carboxylations (involving 2H, 13C, 15N, 17O, 25Mg, and 33S), then the interactions between the telomerases and the DNA change, becoming stronger due to magnetics of fractional, reversible fissing and fusing of the nonprimordial isotopes; so that the telomeres of the DNA open and close like regions in normal telomeres replicating and transcribing nucleic acids; so the nonprimordial telomeres themselves continue to replicate to elongate due to the stronger binding of the nonprimordial telomerases to the nonprimordial telomeres and this causes cancer. External electromagnetic waves and static magnetic fields can be factors affecting such processes for explaining the illusive effects of electric, magnetic and electromagnetic fields on organisms. Thereby cancer develops by random isotopic editing of DNA such that the nonprimordial telomerase bind the nonprimordial telomeres via nonprimordial nonprimordial interactions ; so the telomeres continue to elongate and the DNA continues to replicate. The nonprimordial telomeres thereby fail to shorten and induce apoptosis before the cell reproduces multiple times to transfer epigenetic mutations in DNA, RNA and proteins for cancer habitat. Thereby the theory 1, 2, 3, it is hypothesized that the normal cells have shortened telomeres that stop replicating as they enrich with primordial isotopes: 1H, 12C, 14N, 16O, and 24Mg. But it is further hypothesized that the cancer cells have elongated telomeres with nonprimordial isotopes of 2H, 13C, 15N and 17O that accelerate rather than stop replication due to stronger binding of the telomeres to proteins with nonprimordial isotopes: 2H, 13C, 15N, and 17O. In prior paper 4, the mass spectra of larger pieces of DNA comparable to telomere codon of cancer, white blood cells and red blood cells were compared. In this work, this hypothesis is tested by mass analysis of smaller pieces corresponding to individual nucleotides. Finally, it is hypothesized that drugs and/or food having nonprimordial enriched isotopes like proanthocyanidins can more strongly bind cancer DNA to disrupt the cancer genetics and metabolism for treating cancer.
Method
In order to test some aspects of this hypothesis normal red and white cells and Leukemia cancer cells were obtained and studied in vitro. The DNA from the normal and cancer blood cells were harvested after growth of cancer cells and normal cells in vitro. The DNA was mass analyzed by MALDI mass spectrometry. The mass spectra of the normal and cancer cells were analyzed and compared for isotopic differences. A comparisons of the oligonucleotides of DNA and the oligomeric proanthocyanidins from various fruit and vegetable food sources were done along with the corresponding mass spectra. Similar chemical structures of the proanthocyanidins and DNA oligonucleotides 7 were observed and the anti-cancer effects 8 of proanthocyanidins were reasoned based on exchange of nonprimordial isotopes between the proanthocyanidins and the oligonucleotides.
Results
In Figure 1, the top mass spectrum is for K562 Leukemia Cancer Cells. The middle mass spectrum is for SKW6 Normal Red Blood Cells. The bottom mass spectrum is for tWBC Normal White Blood Cells. Next, the different peak positions for red, white and cancer cells are noted. Table 1, Table 2, Table 3, Table 4, Table 5, Table 6, Table 7, Table 8, Table 9, Table 10 provide details of specific relative intensities at noted peaks. Comparing the spectra, it seems that cancer cells are less abundant at 407.17 Da peak verses more abundant 409.28 Da peak. See Table 1. The peaks at 407 Da - 408 Da correspond to uridine diphosphate or thymidine diphosphate of RNA. The 402 and 403 peaks may be from cytidine diphosphate. The normal cells are more abundant at 407 Da verses 409 Da peaks for more primordial isotopes. Beyond 409 Da to 415 Da peaks (cytosine diphosphate), the cancer cells are less abundant in nonprimordial isotopes, but normal cells are more abundant from 409 Da to 415 Da peaks in nonprimordial isotopes. The peak at 429 Da is from the adenosine diphosphate nucleotide and this nucleotide fragment is found enriched nonprimordials in cancer cells of heavier isotopes in DNA of the cancer cells. See Table 2. The 445 Da peak is from guanosine diphosphate (GTP - PO33–) from RNA seems more abundant in nonprimordial isotopes for white normal cells and red blood cells relative to that in cancer cells. The 444-446 Da peaks distribution in red cells seems heavier than 444-446 Da peaks distribution for white normal cells. See Table 3. The role of 17O may also explain the unusual isotopic content about the 445 Da peak. The 483 and 484 Da peaks correspond to defunctionalized adenosine triphosphate. See Table 4.
Figure 1.Mass Spectra (400 Da to 1000 Da) of DNA from Normal Cells and Cancer Cells
The clumping as by fewer peaks in cancer DNAs is observed and more peaks and finer structure of peaks are observed in normal DNA. The cancer DNA at 483 Da appears to have loss a nonprimordial, more massive functional group from a more massive peak and the normal cells at 484 Da appear appear to have loss a primordial, less massive functional group from a more massive peak. Adenosine triphosphate is observed at peaks of 506 Da and 507 Da; cancer DNA is enriched with nonprimordials at heavier 506 Da peak relative to 503 Da peak for more clumped nonprimordials. See Table 5. Normal DNAs have more enrichment at 503 Da peak with primordials. The peaks at 523-525 Da correspond to guanosine triphosphate and appear enriched with nonprimordials in the cancer DNA. See Table 6. 669 Da and 671 Da peaks are enriched in cancer DNA due to AT monophosphates. See Table 7. The 671 Da peak is enriched in nonprimordial isotopes in cancer relative to 669 Da peak; the white blood cells are enriched in primordial isotopes at 669 Da peak. 675 Da - 676 Da peaks may be AC monophosphates and these peaks reveal cancer is enriched in nonprimordial isotopes relative to white blood cells but these peaks reveal red blood cells are enriched in nonprimordial isotopes at 675 Da peak relative to cancer cells at 673 Da peak. The heavier 675 Da peak in cancer is due to 13C and its 17O. 681 Da - 683 Da peaks may be GC or GT monophosphates. See Table 8. The 680 Da and 681 Da peaks are enriched in primordial isotopes as by the T and C and the cancer is enriched in nonprimordials at 681 Da and 682 Da peaks. See Table 8. 697 Da - 698 Da peaks may be AG monophosphates; the 695 Da peak is enriched in primordial isotopes in white blood cells. See Table 9. The 697 Da peak is enriched in nonprimordial isotopes in the cancer cells. The cancer DNA may have 17O on guanosine and the normal cells may have less guanosine. The peaks at 703 and 709 Da correspond to functionalized AG by OH or NH and the cancer DNA manifest clumped nonprimordials as observed by fewer peaks compared with the finer structure and many peaks of the normal DNA.
Table 1. 407 Da - 409 Da| White Blood Cells | Cancer Cells | Red Blood Cells | ||||||
| 400.144 | 0.039 | 0.097 | 402.221 | 0.063 | 0.062 | 400.128 | 0.016 | 0.099 |
| 400.342 | 0.023 | 0.089 | 403.278 | 0.037 | 0.086 | 400.329 | 0.013 | 0.122 |
| 401.138 | 0.045 | 0.114 | 405.146 | 0.045 | 0.06 | 401.117 | 0.014 | 0.086 |
| 402.231 | 0.093 | 0.103 | 407.169 | 0.248 | 0.059 | 402.222 | 0.031 | 0.109 |
| 403.124 | 0.026 | 0.076 | 408.184 | 0.05 | 0.049 | 403.137 | 0.012 | 0.058 |
| 403.282 | 0.050 | 0.116 | 409.283 | 0.133 | 0.063 | 403.271 | 0.023 | 0.122 |
| 404.269 | 0.050 | 0.117 | 404.268 | 0.017 | 0.142 | |||
| 405.171 | 0.043 | 0.179 | 405.16 | 0.020 | 0.209 | |||
| 406.166 | 0.020 | 0.100 | 406.198 | 0.011 | 0.09 | |||
| 406.326 | 0.018 | 0.112 | 406.322 | 0.011 | 0.100 | |||
| 407.18 | 0.296 | 0.096 | 407.169 | 0.101 | 0.091 | |||
| 408.185 | 0.043 | 0.089 | 407.321 | 0.015 | 0.101 | |||
| 408.315 | 0.018 | 0.115 | 408.174 | 0.018 | 0.090 | |||
| 408.341 | 0.019 | 0.098 | ||||||
| White Blood Cells | Cancer Cells | Red Blood Cells | ||||||
| 425.229 | 0.035 | 0.169 | 425.283 | 0.044 | 0.075 | 425.268 | 0.024 | 0.135 |
| 426.292 | 0.018 | 0.199 | 429.153 | 0.248 | 0.049 | 426.277 | 0.011 | 0.202 |
| 427.236 | 0.032 | 0.167 | 430.161 | 0.051 | 0.049 | 427.25 | 0.011 | 0.089 |
| 429.164 | 0.242 | 0.099 | 430.161 | 0.051 | 0.049 | 428.162 | 0.010 | 0.112 |
| 430.173 | 0.032 | 0.113 | 436.114 | 0.040 | 0.069 | 429.151 | 0.110 | 0.096 |
| 430.346 | 0.018 | 0.012 | heavier cancer | 430.352 | 0.011 | 0.109 | ||
| 431.187 | 0.019 | 0.120 | 431.15 | 0.011 | 0.100 | |||
| 436.116 | 0.062 | 0.093 | 436.108 | 0.018 | 0.097 | |||
| White Blood Cells | Cancer Cells | Red Blood Cells | ||||||
| 442.234 | 0.021 | 0.212 | 442.255 | 0.012 | 0.239 | |||
| 443.336 | 0.034 | 0.140 | 439.119 | 0.050 | 0.032 | 443.336 | 0.014 | 0.135 |
| 444.344 | 0.015 | 0.112 | 445.101 | 0.100 | 0.061 | 444.344 | 0.013 | 0.08 |
| 445.118 | 0.193 | 0.095 | 455.302 | 0.044 | 0.080 | 445.118 | 0.069 | 0.095 |
| 445.298 | 0.067 | 0.102 | 446.117 | 0.016 | 0.108 | |||
| 446.117 | 0.031 | 0.14 | ||||||
| White Blood Cells | Cancer Cells | Red Blood Cells | ||||||
| 480.318 | 0.020 | 0.214 | ||||||
| 481.236 | 0.022 | 0.105 | 481.358 | 0.012 | 0.125 | |||
| 482.078 | 0.035 | 0.092 | 482.199 | 0.015 | 0.088 | |||
| 483.073 | 0.050 | 0.095 | 483.065 | 0.02 | 0.096 | |||
| 483.231 | 0.035 | 0.071 | 483.217 | 0.015 | 0.075 | |||
| 483.372 | 0.050 | 0.097 | 483.391 | 0.074 | 0.069 | 483.37 | 0.03 | 0.096 (R hv vs W) |
| 484.044 | 0.022 | 0.097 | 484.041 | 0.027 | 0.101 | |||
| 484.258 | 0.019 | 0.081 | ||||||
| 484.378 | 0.057 | 0.097 | 484.381 | 0.052 | 0.087 | 484.377 | 0.031 | 0.104 (R hv vs W) |
| 485.047 | 0.028 | 0.103 | 485.046 | 0.027 | 0.098 | |||
| 485.246 | 0.033 | 0.085 | 485.472 | 0.024 | 0.178 | |||
| 485.374 | 0.037 | 0.105 | 486.048 | 0.015 | 0.099 | |||
| 486.257 | 0.027 | 0.214 | 486.38 | 0.011 | 0.181 | |||
| 487.287 | 0.296 | 0.101 | 487.27 | 0.165 | 0.055 | 487.045 | 0.01 | 0.103 (R Lt vs W) |
| 488.291 | 0.084 | 0.113 | 488.276 | 0.049 | 0.058 | 487.279 | 0.067 | 0.099 (W peak) |
| 489.072 | 0.037 | 0.116 | 489.047 | 0.042 | 0.066 | 488.038 | 0.02 | 0.097 |
| 488.256 | 0.024 | 0.197 | ||||||
| 489.051 | 0.047 | 0.101 (R hv W) | ||||||
| White Blood Cells | Cancer Cells | Red Blood Cells | ||||||
| 498.385 | 0.017 | 0.098 | 499.366 | 0.043 | 0.083 | 499.703 | 0.022 | 0.099 |
| 499.254 | 0.029 | 0.072 | 500.367 | 0.044 | 0.089 | 500.0270 | 0.023 | 0.102 |
| 499.367 | 0.065 | 0.101 | 501.242 | 0.092 | 0.055 | 500.388 | 0.020 | 0.104 |
| 500.257 | 0.021 | 0.074 | 503.254 | 0.043 | 0.083 | 501.222 | 0.032 | 0.099 |
| 500.390 | 0.058 | 0.100 | 505.263 | 0.463 | 0.064 | 501.386 | 0.015 | 0.109 |
| 501.241 | 0.121 | 0.099 | 506.264 | 0.113 | 0.070 | 502.254 | 0.013 | 0.091 |
| 501.390 | 0.037 | 0.107 | 507.266 | 0.065 | 0.080 | 502.378 | 0.018 | 0.097 |
| 502.251 | 0.042 | 0.095 | 515.362 | 0.051 | 0.064 | 503.250 | 0.056 | 0.112 |
| 502.386 | 0.031 | 0.096 | Cancer is lighter at 506 Da | 504.251 | 0.019 | 0.102 | ||
| 503.260 | 0.190 | 0.109 | No peaks in at 503 Da | 504.385 | 0.012 | 0.117 | ||
| 504.265 | 0.067 | 0.119 | 505.261 | 0.161 | 0.101 | |||
| 505.068 | 0.017 | 0.115 | 505.393 | 0.020 | 0.07 | |||
| 505.272 | 0.530 | 0.106 | 506.260 | 0.046 | 0.103 | |||
| 506.278 | 0.138 | 0.111 | 507.277 | 0.028 | 0.100 | |||
| 507.292 | 0.093 | 0.124 | 507.388 | 0.018 | 0.096 | |||
| 508.295 | 0.028 | 0.170 | 508.328 | 0.011 | 0.205 | |||
| 509.283 | 0.024 | 0.246 | 509.408 | 0.014 | 0.124 | |||
| White Blood Cells | Cancer Cells | Red Blood Cells | ||||||
| 521.332 | 0.024 | 0.216 | 522.289 | 0.050 | 0.047 | 521.371 | 0.012 | 0.113 |
| 522.310 | 0.056 | 0.129 | 525.418 | 0.036 | 0.116 | 522.305 | 0.018 | 0.136 |
| 523.233 | 0.047 | 0.128 | 527.238 | 0.055 | 0.068 | 523.220 | 0.015 | 0.111 |
| 524.234 | 0.023 | 0.108 | 529.240 | 0.035 | 0.081 | 523.403 | 0.011 | 0.083 |
| 525.247 | 0.034 | 0.095 | 524.208 | 0.010 | 0.113 | |||
| 525.400 | 0.033 | 0.119 | 524.403 | 0.011 | 0.110 | |||
| 526.266 | 0.024 | 0.256 | 525.254 | 0.014 | 0.098 | |||
| 527.252 | 0.080 | 0.099 | 525.388 | 0.022 | 0.112 | |||
| 527.401 | 0.030 | 0.117 | 526.389 | 0.013 | 0.122 | |||
| 528.265 | 0.029 | 0.084 | 527.246 | 0.026 | 0.102 | |||
| 528.401 | 0.066 | 0.097 | 527.400 | 0.011 | 0.126 | |||
| 529.265 | 0.048 | 0.097 | 528.249 | 0.011 | 0.088 | |||
| 529.390 | 0.044 | 0.105 | 528.396 | 0.020 | 0.104 | |||
| 530.334 | 0.032 | 0.227 | 529.255 | 0.019 | 0.098 | |||
| 529.398 | 0.014 | 0.097 | ||||||
| 530.333 | 0.013 | 0.240 | ||||||
| White Blood Cells | Cancer Cells | Red Blood Cells | ||||||
| 666.179 | 0.021 | 0.122 | 665.136 | 0.077 | 0.041 | 666.178 | 0.011 | 0.138 |
| 666.428 | 0.016 | 0.150 | 665.231 | 0.041 | 0.066 | 667.247 | 0.019 | 0.121 |
| 667.299 | 0.017 | 0.156 | 669.258 | 0.098 | 0.083 | 668.234 | 0.025 | 0.115 |
| 667.369 | 0.016 | 0.041 | 670.264 | 0.046 | 0.098 | 669.231 | 0.100 | 0.115 |
| 669.273 | 0.116 | 0.114 | 671.287 | 0.091 | 0.064 | 669.423 | 0.010 | 0.129 |
| 669.431 | 0.025 | 0.093 | 672.277 | 0.052 | 0.064 | 670.221 | 0.051 | 0.117 |
| 670.274 | 0.055 | 0.113 | 673.297 | 0.186 | 0.058 | 671.256 | 0.039 | 0.124 |
| 670.443 | 0.020 | 0.117 | 674.298 | 0.081 | 0.057 | 671.449 | 0.010 | 0.140 |
| 671.290 | 0.100 | 0.124 | 675.217 | 0.069 | 0.051 | 672.248 | 0.022 | 0.118 |
| 672.298 | 0.040 | 0.150 | 675.309 | 0.041 | 0.106 | 672.725 | 0.010 | 0.120 |
| 673.307 | 0.221 | 0.121 | 676.211 | 0.039 | 0.070 | 673.293 | 0.061 | 0.129 |
| 673.715 | 0.017 | 0.104 | 673.465 | 0.012 | 0.126 | |||
| 674.102 | 0.031 | 0.119 | 674.160 | 0.015 | 0.084 | |||
| 674.311 | 0.085 | 0.122 | 674.307 | 0.026 | 0.105 | |||
| 675.100 | 0.039 | 0.111 | 675.212 | 0.079 | 0.116 | |||
| 675.317 | 0.048 | 0.139 | 675.351 | 0.011 | 0.143 | |||
| 676.285 | 0.022 | 0.118 | 676.205 | 0.046 | 0.116 | |||
| 676.436 | 0.018 | 0.103 | ||||||
| White Blood Cells | Cancer Cells | Red Blood Cells | ||||||
| 678.310 | 0.038 | 0.158 | 675.309 | 0.041 | 0.106 | 676.205 | 0.046 | 0.116 |
| 678.390 | 0.021 | 0.113 | 676.211 | 0.039 | 0.070 | 677.221 | 0.017 | 0.115 |
| 679.283 | 0.019 | 0.095 | 681.092 | 0.038 | 0.051 | 677.380 | 0.013 | 0.152 |
| 679.465 | 0.017 | 0.133 | 681.257 | 0.152 | 0.060 | 678.244 | 0.011 | 0.112 |
| 680.440 | 0.016 | 0.115 | 682.246 | 0.125 | 0.073 | 679.224 | 0.012 | 0.125 |
| 681.100 | 0.019 | 0.124 | 683.250 | 0.037 | 0.065 | 680.215 | 0.012 | 0.142 |
| 681.263 | 0.054 | 0.111 | 687.114 | 0.140 | 0.055 | 681.090 | 0.020 | 0.094 |
| 681.447 | 0.025 | 0.090 | 681.253 | 0.113 | 0.113 | |||
| 685.442 | 0.016 | 0.129 | 682.241 | 0.126 | 0.116 | |||
| 686.430 | 0.026 | 0.137 | 683.246 | 0.033 | 0.112 | |||
| 0.019 | 0.133 | 684.249 | 0.012 | 0.117 | ||||
| 685.494 | 0.011 | 0.156 | ||||||
| 687.113 | 0.037 | 0.116 | ||||||
| White Blood Cells | Cancer Cells | Red Blood Cells | ||||||
| 692.250 | 0.023 | 0.129 | 687.114 | 0.140 | 0.055 | 692.207 | 0.018 | 0.120 |
| 693.263 | 0.023 | 0.114 | 691.242 | 0.045 | 0.094 | 693.218 | 0.011 | 0.130 |
| 695.283 | 0.065 | 0.117 | 695.270 | 0.046 | 0.059 | 695.265 | 0.021 | 0.131 |
| 695.460 | 0.016 | 0.146 | 697.288 | 0.042 | 0.024 | 696.246 | 0.015 | 0.220 |
| 696.081 | 0.028 | 0.115 | 703.075 | 0.039 | 0.044 | 697.044 | 0.010 | 0.075 |
| 696.283 | 0.031 | 0.115 | 697.211 | 0.042 | 0.120 | |||
| 697.082 | 0.039 | 0.116 | 698.206 | 0.023 | 0.132 | |||
| 697.293 | 0.032 | 0.148 | 699.270 | 0.010 | 0.208 | |||
| 698.434 | 0.018 | 0.110 | 699.508 | 0.010 | 0.139 | |||
| 699.319 | 0.026 | 0.232 | ||||||
| White Blood Cells | Cancer Cells | Red Blood Cells | ||||||
| 703.413 | 0.016 | 0.130 | 703.075 | 0.039 | 0.044 | 703.066 | 0.018 | 0.100 |
| 704.242 | 0.016 | 0.106 | 709.088 | 0.106 | 0.063 | 703.238 | 0.023 | 0.103 |
| 704.430 | 0.017 | 0.140 | 719.039 | 0.036 | 0.052 | 704.233 | 0.024 | 0.113 |
| 705.438 | 0.020 | 0.136 | 705.238 | 0.010 | 0.113 | |||
| 707.254 | 0.018 | 0.113 | 709.089 | 0.036 | 0.118 | |||
| 709.095 | 0.083 | 0.117 | 713.184 | 0.021 | 0.117 | |||
| 710.094 | 0.021 | 0.107 | 714.167 | 0.019 | 0.114 | |||
| 712.439 | 0.018 | 0.113 | ||||||
| 714.292 | 0.023 | 0.168 | ||||||
Discussion
General
The observed mass spectra of the DNA of normal and cancer cells and the displacements of the peaks in the range 400 Da to 1000 Da may be interpreted on the basis of the fragmentations of the DNA into nucleosides, nucleotides and oligonucleotides during MALDI mass analysis process with varying characteristic isotopic compositions of 2D/1H, 13C/12C, 15N/14N, 17O/16O and/or 25Mg/24Mg within the fragments. The C to T → U and A → G has methylations, dehydrations, deaminations and hydrations of rings of aromatics, purines, pyrimidines and ribose rings and phosphate groups as isotopically exchanged functional groups. On the basis of these varying isotopic compositions of the DNA in cancer and normal cells, the differing fragmentation patterns of the DNA can be reasoned. The varying isotopic contents can also be reasoned by different interactions, formations, replications, transcriptions, and translations of these nucleic acids in normal cells verses cancer cells.
Discussion of 13CH3 for Enriched Thymidine
The 400-409 Da peaks may be U, T or C. The interconversions may be due to the nonprimordials so as to cause enrichments and depletions. See Figure 2. The T is at 402 Da, the C is at 403 Da and the U is at 404 Da. The U can → T by dehydroxylation and methylation and vice versa. And the T can → C by dehydroxylation and amination and vice versa. By the prior theory of Little 1, 2, 3, hydroxylation is critical for both these interconversions as the OH is strong nucleophile and less subject to difficulty rehybridization dynamics due to more electron ××× electron interactions about O nuclei and the attached p+. The 17OH accelerates both the bond breakages for methylations and aminations. This theory determines important 17OH2 and 15NH3 nanosolutions 1, 2, 3 inside cancer cells such that the nanosolvent 17OH2 and 15NH3 weaken bonds in the nucleotides to accelerate 13CH3 nucleophillic replacement in the aromatics for kinetics of the mechanism 1, 2, 3. Typically, aromatics are so stable that they are more difficult for nucleophilic aromatic substitutions, but the negative nuclear magnetic moments (NMMs) of 17O and 15N lower the activation energies for accelerated substitutions on the nucleophiles 2, 3. The 1H on 17O and 15N modulates the negative NMMs of 17O and 15N for perturbing the covalent bonds in the nucleotides via oscillating e- e- pairs of the bases; strong electron --- electron interactions in valance of O further facilitate dynamics for lability. The 15NH3 and 17OH are by their negative NMMs softer bases and better leaving on the basis of this theory; so they explain the large massive loss from mass spectra of cancer DNA. These results and predictions of this theory 1, 2, 3 are consistent with prior experimental observations of NH3 recycling by breast cancer cells. But researchers have not experimentally explained 17OH2 in cancer cells 1, 2, 3. But this theory predicts large anomalous dynamics of 17OH2 in cancer cells 2, 3. The methylations are faster as 13C has positive NMM. The negative NMM of 15N may alter its amination of reaction centers. Dense nonzero NMMs may deaccelerate the 15NH2 deamination.
Figure 2.Pyrimidine Nucleotides
The nonprimordials in U may cause it to manifest 409 Da peak in the cancer and the enrichments of nonprimordials and clumping in the cancer DNA. Such nonprimordials in cancer in U may accelerate its conversion to T by accelerating dehydroxylations of ribose and accelerating methylations of pyrimidine by :17OH2 complexation and :15NH3 complexation of U from surrounding nano-NH3 and H2O nanosolution. The 16OH in the ribose of cancer’s U more rapidly fragments its bond to the ribose due to the null NMM of the 16OH and the 17OH rapidly replaces 16OH, but during mass analysis 17OH may be fragmenting from T nucleotides of cancer DNA to explain the spectra. The 13CH3 is better, stronger nucleophile and more rapidly attacks the pyrimidine due to the positive NMM of 13C of 13CH3. The nonprimordials at 409 Da peak may thereby more rapidly form the T at 402 Da peak with enriched nonprimordials at 402 Da in cancer. It is important to note further that this theory determines greater solubilities and greater complexations of 13CH3 by nanowaters of :17OH2 and :15NH3 relative to complexations and solubilities of primordial 12CH3. The cancer DNA is heavier at 403 Da and indeed the clumping of nonprimordials in the formed T has even greater intensity in cancer DNA at 405 Da (vs 403 Da) relative to normal cell at 403 Da vs 405 Da. The methylations of the U and dehydroxylations are expected by prior theory 1, 2, 3 to have enriched nonprimordials in the T in cancer DNA. (So in general the 13CH3 is a stronger better nucleophile and stronger base than 12CH3 due the harder basicity 13CH3 relative to 12CH3 2, 3. So now in general 17OH (15NH2) is a better leaving group than 16OH (14NH2) and a weaker base than 16OH (14NH2) due to the greater polarizability of 17OH (15NH2) to 16OH (14NH2) 2, 3.
Discussion of Interconversion of Pyrimidines (C, T and U)
The T may convert to C by deaminations and hydroxylations and demethylations. So in cancer the nonprimordials may accelerate the deaminations of T as 15NH2 is a poor leaving group relative to 14NH2. See Figure 2. But the hydroxylations may be more in cancer (as 17OH is a betterer nucleophile than the 16OH) relative to deaminations in cancer DNA for kinetics and thermodynamic reasons. But the: OH2 is stronger base than :NH2 and kinetically electronic rehybridizations are more labile in OH due to greater electron density about the O nucleus relative to N nucleus 2, 3. Thermodynamically the O-H bonds are stronger than N-H bond due to the larger effective nuclear charge of O. The: O may be stronger base as its nucleus pulls: e- e- pair more strongly with denseness than : N. So denser e- e- pull proton electro-statically. The zero NMM of 16O and positive NMM of 14N cause magnetic pull of e- e- by: 14N, but the electrostatic extra p+ in 16O nucleus pulls more the e- e- pair for its greater basicity. But as 16O goto 17O the null NMM goes to negative NMM for larger more polarizable e- e- cloud and also as 14N goes to 14N the positive NMM goes to negative NMM for even larger relative change in e- e- cloud size and polarizability. So both 17O and 15N are weaker bases but softer bases relative to 16O and 14N. The nuclei magnetically polarize the e- e- about 17O and 15N and the interactions with the p+ is magnetic. And the interaction changes abruptly as temperature increases, pressure increases and electric and magnetic fields stimulate so depending on conditions stronger magnetic interactions can cause 15N and 17O to be stronger bases and change their nucleophilicities.
In cancer, the demethylations may be slower than in normal cells as 13CH3 of the cancer may be poor leaving group. So the cancer may less transform its T* to C* relative to the normal cells so the peaks for C* (403 Da) in cancer should be depleted in nonprimordials isotopes. The 402 Da peak for C is observed depleted in primordials in the cancer relative to the observed 402 Da in the normal DNA, revealing more primordial in normal DNA. In fact, the normal cells show peaks at both odd and even masses 402, 403, 404, 405, 406, 407, 408, 409 and 410 Da as the T* at 403 Da shows nonprimordials for odd masses and the T at 402 Da for less clumping of even masses of nonprimordial isotopes in the normal cells. But the cancer DNA shows only odd peaks 403 Da, 405 Da, 407 Da and 409 Da for the formation of T* (403 Da), but no formations of C* (404 Da) are observed in the cancer as the U* → T*. But T* does not → C* in cancer. The data thereby reveal depleted C* in the cancer with enriched T*. But all these transformations occur in normal cells. It may be that during cell division cancer nuclei have too much thymidine (T) and too little cytidine (C) as thymidine accumulates in normal cells to cause the normal cells to transform to cancer cells.
The depletion of the primordials in cancer DNA at 409 Da peak for UTP and enrichment of primordials in cancer DNA at 407 Da peak may be reasoned by cytidine becoming more rapidly methylated with 13CH3 relative to 12CH3 during cancer genesis so that the cytidine may transform to uridine and thymidine for altering the (13CH3) isotopic compositions of C, T and U in the cancer cells as the nonprimordial C in cancer more rapidly functionalizes and defunctionalizes to enrich T and U with nonprimordials (13C). It may be that during the mass spectra under the electromagnetism the cancerous DNA less readily fragments (under the strong electromagnetism of the mass spectrometer) near dense regions of nonprimordial kernels; so the cancer DNA has fragments with nonprimordial enrichment at 409 Da peak and enrichment of primordials at 407 Da peak. The 13CH3 is a stronger nucleophile by this theory so it more rapidly attacks C in the 15NH3, 17OH2 solutes about DNA in nanosolutions in cancer cells 2, 3. This theory thereby predicts and explains more rapid methylation of C in DNA of cancer. Moreover, this theory further discloses the more rapid conversion of C* to U as the surrounding nanowater in cancer cells complexes the 16O and 14N in the C to accelerate conversions in C* (methylated) C and to then accelerate the deaminations of pyrimidine of C* for loss of 14N and replacements by OH to form U*. Thereby C is not only by this theory 1, 2, 3 prevented from forming from T*. The C in cancer is accelerated to form U*. It may also be possible that 17O is enriching in the phosphorate group of the nucleotide. So the cancer is heavier beyond 409 Da to 415 Da peaks due to 17O on phosphates and varying protonation of 17O on 31PO3- and 17O and 15N on the purines of guanosine diphosphate. Normal cells have 16O on 31PO3 with stronger protonation and 16O and 14N on purines of guanosine, but cancer DNA may accumulate 17O in 31PO3.
Clumping of Nonprimordials in Cancer DNA and Enriched Adenosine from Guanosine
The 429 Da peak may be enriched in nonprimordials in cancer due to adenosine diphosphate and its formation from guanosine diphosphate by dehydrations and deaminations of G; and aminating the intermediate may thereby result from faster functionalizations and defunctionalizations of ribose and pyrimidine rings by methylations and deaminations for thymidines having 13CH3. See Figure 3. The normal cells have more local peaks about 429 Da relative to cancer 429 Da peak as cancer has more clumped nonprimordials. So the 429 Da nucleotide with nonprimoridial 17O in cancer DNA rapidly loses functional group; so the 429 Da peak is less present, but the fragments in the cancer having primordial 16O show larger peaks as they fragment less by loss of their 16O. 17O is more labile than 16O. The formation of the 429 Da from the 445 Da for G → A involves deaminations and dehydrations of G then aminations to A. The cancer cells have nanowater with 17OH2 and 13NH3 to complex the OH and NH2 of G in cancer to convert to G* with 15N and 17O replacement in cancer G*. The 15N and 17O more readily undergo nucleophilic aromatic substitutions by 14N to form the A from the G by this theory 1, 2, 3 to explain the data. The clumping of nonprimordials in cancer DNA and G nucleotide may accelerate the deaminations and dehydroxylations of G as cancer has 15NH2 and 17OH, which are poor nucleophiles and good leaving groups due to their negative NMMs. The 15NH2 is a stronger nucleophile than 17OH as 15N has a less negative NMM than 17O; so 15NH2 is expected to be harder than 17OH. 15NH2 is expected to be a better nucleophile than 17OH for replacing 17OH of G* to form 15NH2 of A*, so the 15NH2 is harder base and should attack the purine rings faster than 17OH weaker nucleophile. Thereby cancer DNA should readily transform G*→ A* for unusual mutations. It may be that cancer cells show excess of adenosine and deficiencies of guanosine so this may reflect in anomalous RNA transcriptions and protein translations in cancer anabolism. Red blood cells show similar isotopic distributions relative to normal cells as the thymidine and cytidine may not functionalize their ring with 17O as 17O defunctionalizes from guanosine.
Clumping of Nonprimordials in Cancer DNA and Enriched Guanosine → Adenosine Diphosphate
The unusual enrichments in the 445 Da peak of cancer with nonprimordials (relative to 443 Da for primordial G) may be explained by the A* in the 429 Da peak of A* as the A may reversibly undergo uphill slower process of deaminations, hydroxylations and aminations to form the G at 445 Da peaks. Thereby the clumped nonprimordials in G more accelerate the loss of 15NH and the gain of 15N and 17O to form A, relative to primordials in A of normal cells to form the nonprimordial G in the cancer. So the peak at 445 Da peak is nonprimordial enriched in cancer DNA as the nonprimodials compose A, leaving the nonprimordial enriched G at 445 Da peak rather than 443 Da. See Figure 3. Cancer DNA at 445 Da is heavier with nonprimordials relative to normal white cells DNA. Vice versa in the cancer cell the G* may transform readily due to its clumped nonprimordial isotopes transform to A*. The dehydroxylations and deaminations and aminations of the G* in cancer DNA are accelerated due to the negative NMMs of the 15N and 17O for alterations of leaving ability in the dehydroxylations and deaminations. But the aminations to form the final A* is expected in cancer DNA as the 15NH2 is a weaker base and weaker nucleophile than 17OH due to the harder basicity of 15NH2 relative to 17OH and due to the less negative NMM of 15NH2 relative to more negative NMM of 17OH-. The hydrogens also help harden the 15NH2- as there are more H in 15NH2 relative to 17OH. Stronger internal magnetism to stabilize 14NH3 verses 15NH3 has greater stability of 15NH3 so 14NH3 goto N2 + H2 faster and stronger bonds. 15N triple bond is less stable. 14N triple bond may be easier to break. So breaking 15N2 may be easier than breaking 14N2 and the stability of 15N-R in organic compounds may be greater than 14N-R toward N2 formations.
Uridine, Cytidine and Thymidine Triphosphides and Nonprimordial Clumps Block T → C in Cancer DNA
So the heavier U (484 Da), T (482 Da) and/or C (483 Da) in cancer couple by chemical transformations to 502 Da of U* (methylated U) in cancer by methylations (13CH3, 16Da); by U* dehydroxylating to form T* (with 13CH3, 16Da); by U* dehydroxylating (17OH, 18Da) and aminating (15NH2, 17 Da) to form C*. See Figure 2. Such many possible dynamics manifest nonprimordial accelerated functionalizations and defunctionalizations in cancer cells relative to normal cells and the resulting nonprimordial induced chemical transformations of U → U*, U* → T* and/or C* → U* for new mechanisms of mutations of DNA and RNA as here disclosed as by not only by changing isomeric connectivity along chains but also by interchemically converting nucleotides of U, T and C! Such complex inter-chemical conversions are observed in the mass spectra of the cancer relative to the normal cells. So the normal cells have finer peaks in this range 400 - 409 Da. The cancer DNA less fragments to form U, T, and C and have less fine spectra due to nonprimoridials clumping from 400 to 409 Da. The nonprimordials (13CH3) in the cancer nucleotides may cause less fragmenting of cancer DNA for fewer of these peaks from 400 - 409 Da. The normal cells have random methylations, the random methylations of C and random 13CH3 in C, T, and U can lead to such fine peaks in the normal cells. But the cancer cells have nonrandom, clumping of 13CH3 and 15NH2 and 17OH and the 13CH3 causes stronger binding of the cancer DNA for less fragmentations under electromagnetic fields in mass spectrometer. There is more 483 Da in cancer DNA and there is more 484 Da in normal DNA, there is more T* in cancer DNA and more C* in normal DNA. These trends for cytidine, uridine and thymidine triphosphates are consistent with the peaks at 400-409 Da for the diphosphates as the diphosphates also revealed less T to C for cancer DNA. The T* → C* conversions in cancer would involve the dehydroxylations of pyrimidine and hydroxylations of ribose and the aminations of thepyrimidine. The negative NMMs of the 17OH and 15NH2 in cancer make this less likely. As the negative NMM of 15N and 17O make 15NH2 better nucleophiles for the conversions of the pyrimidine to C* in cancer DNA but instabilities. The cancer DNA thereby less expresses C* at 483 Da.
Dehydration of Adenosine Triphosphate and Suppresion by Clumping 13CH3
The 487 Da is from the dehydroxylations (17OH or 18OH 17O2D or 18O2D of 18 Da to 20 Da) of ribose in adenosine triphosphate at 507 Da. See Figure 3. In normal cells, the dehydroxylations are more than in cancer cells as the cancer cells have more 13C in the ribose, which bind the 17OH more strongly. So cancer at 507 Da should be heavier. But cancer is observed not to be heavier at 507 Da as the 507 Da is coupled to 523 – 525 Da by dehydroxylations. So 523 Da to 525 Da of G* in cancer losses 17O (rather than 16O) to cause less massive peaks at 482 Da. Cancer hydroxylates better if it is heavier (clumped with 13C or 15NH3), so the 507 Da peak in cancer lacks heavier nonprimordials as they are loss of 17OH from 523 – 525 Da to G*. So they are missing 507 Da peak A* at 525 Da peak in G*. This conversion of nonprimordial G* to nonprimordial A* in the cancer DNA is expected as the G* to A* involves the deaminations, hydroxylations and aminations of the purine. The cancer having clumped nonprimordials may accelerate this as the 15NH2 and 17OH in the cancer DNA are weaker nucleophiles (due to their negative NMMs) relative to 14NH2 and 16OH. But in principle 15NH2 and 17OH should be poorer entering groups due to their negative NMMs but ring 15N can pull in the 15NH2 and 17OH nucleophiles. It is observed that cancer is heavier at 525 Da G* relative to normal cells being lighter at 523 Da. As the 523-525 Da is guanosine triphosphate and the 17O on the guanosine triphosphate stabilize the 17O and the 13CH3 for less massive peaks at 505 Da and less massive peaks 487 Da in the cancer DNA samples due to losses of heavier 17O and 13C, respectively. So this is general principle when 17O is active in fragmenting, the daughter peaks are enriched in less massive than peaks in primordial normal DNA. When 13CH3 is active in the fragmenting the daughter peaks are enriched in nonprimordials as the 13CH3 fragments stabilize by 13CH3 with consequent heavier daughter peaks. The nonprimordials in the 17OH destabilize and the 13CH3 stabilizes primordials. So the enrichments in the daughter by primordials are actually due to lack of instability or less fragmenting of nonprimordial than primordials.
All + or – NMMs activate bond breaking. All – NMMs have faster kinetics of bond breaking to cause new effects relative to all + NMMs. Faster kinetics can lead to different product distributions and breaking stronger bonds. All negative NMMs may break C-C, C-O, C-H, O-H bonds and all positive break C=C ⟷ C-C The activated state may then better bond back together by + NMMs + +NMMs with faster rates and with more thermodynamic stability. Such may be more discerned under conditions of high temperature, strong electric fields, strong magnetic fields and/or high pressures. The π bonds in DNA and RNA makes it easier to alter by NMMs, explaining why reproductions, transcriptions and translations are more affected by nonprimodials relative to glycolysis and Kreb cycle. Amino acids having π bonds like tyrosine and phenyl alanine (C=C) may more easily be affected by nonprimordials. Aspartate and glutamate have carbonyl side groups with resonating pi bonds. But amino acids have C=N and C=O and C=C but not aromatic. Carbonyls have resonating C=O.
Adenosine Triphosphate Form from Guanosine Triphosphate
The 506 Da and 507 Da peaks can also be explained on the basis of their A contents. It can be that guanosine triphosphate at 523 Da peak loses 17O to form 507 Da peak {which corresponds to adenosine triphosphate} and the clumped nonprimordials help loss of 17O to explain the patterns. G → A. See Figure 3. The nonprimordial G at 525 Da peak more rapidly loses 17O to produce more than 50% greater loss than 16O is lost to produce 507 Da in the cancer. Thereby here it is proposed that nonprimordial isotopes epigenetically alter nucleic acids in cancer by causing G →A. The 523 Da peak may involve transformations between A and G with a surrounding peak; so that in cancer there is peak enrich in primordial isotopes. A → G by hydroxylations, deaminations, and aminations. G → A by dehydroxyations, deaminations and aminations. Ammonia in tumor can encourage aminating and deaminating G and A, and also induce C → T.
Heaviness of AT in Cancer DNA
The AT fragment associated with the 669 Da to 671 Da peaks and T in AT may be the reason the cancer DNA is enriched in nonprimordial isotopes as the T may form from 13CH3 methylations of cytosine and the cytosine may undergo deaminations and dehydroxylations or the C may → U by deaminations and hydroxylations under acidic conditions as in altered nucleuses (isotopic replacements) as nucleuses are more basic than cytoplasma. The more basic nucleus in cells stabilize T and U as T is more basic and nonpolar relative to U. So in cytoplasma, the T → U as the more acidic cytoplasma can push out 13CH3. It is quite interesting that AT are detected as in cancer AT are thought enriched and GC are thought deficient in cancer. Again in the cancer the accumulations of nonprimordial T* are observed as the T* cannot (due to clumped nonprimordials) convert to C* as the conversion of T* would require demethylations (loss of 13CH3). The 13CH3 is a strong base and good nucleophile and the cancer cells cannot as well lose 13CH3. The heavier 675 Da peak in cancer is due to 13C and its 17O.
The 680 Da and 681 Da peaks may be explained by isotopic distributions in GC or GT. The 680 Da and 681 Da peaks of normal cells are enriched in primordial isotopes as by the T and C having more 12CH3 and 14N2; but the cancer DNA is enriched in nonprimordials at 681 and 682 Da peaks due to the isotopic clumping of nonprimordials to enrich the 13C methylation of C to form 13CH3 in T* also having 15N. There is more GT in cancer than normal cells. There is more GT in cancer than AT. GT has stronger binding due to the 3 hydrogen bonds relative to only 2 hydrogen bonds in AC. G is deficient, so why so much TG? Although deficient G binds strongly to T. Again the enrichments of 13CH3 in T* in nonprimordial cancer is detected and the inability to convert T* to C* in the cancer increases T*. Red Blood Cells are enriched at 682 Da peak relative to cancer at 681 Da peak and this could be due to 17O in G, C and T in the red blood cells as the red blood cells couple to air for ready oxygenation. It may be possible to relate cancer to 17O from the air as well as 17O in the water. So the blood can accumulate 17O from 17O2 and H217O and 13C17O. The red blood cells are different from white blood cells. The red blood cells may be a basis for the cancer spreading the 17O to normal cells.
GA and Loss of G in Cancer DNA
The unusual enrichment of primordial isotopes in cancer AG at 695 Da and 697 Da peaks may be reasoned on basis of G content in GA and the cancer may have 17O and 15N on guanosine and many normal cells have less 17O and 15N on guanosine. There is observed that there is less GA in cancer DNA than GT or GA fragments less than GT. Less observed GT is consistent with the discovery of transforming G to A in cancer genesis by this work. But the observed greater 695 Da relative to 697 Da in cancer may be explained by this theory. So the 17O is more rapidly lost from guanosine of cancer DNA relative to less lost of 16OH from guanosine for the greater 695 Da peak relative to 697 Da peak for cancer. The 695 Da may be coupled thereby to 695 Da+ 14 Da = 709 Da peak or the 695 Da + 9 Da = 703 Da. This 703 Da peak should be enriched in clumped nonprimordials in the cancer as by loss of O2- from G or A. The 14 Da may be loss of 14 Da or NH2 – from G or A. The cancer DNA shows both 703 and 709 Da peaks and manifest this clumping. But the normal cells do not show such peaks at 703 Da and show a small peak at 709 Da in support of this reasoning. The guanosine may be more reactive due to 17O relative to 15N as the 17OH is stronger nucleophile than the 15NH3; and NH3 is less abundant in normal cells! It seems in general 17O helps decompositions and fragmentations. The 17OH2 and 15NH3 in surrounding nano-water in cancer cells may accelerate exchange of 12NH2 and 16OH by 13NH2 and 17OH. Scientists have not measured 17O in mass spectra and NMR enough to see this effect of 17O as determined in this work. Most prior work on O has focused on 16O and 18O. The complexations of this biomolecules by 17OH and 15NH2 cause softening of the bonds for faster substitution and replacement reactions due to the negative NMMs of 17O and 15N.
So in general where 13CH3 reactions are accelerated in cancer, the methylation consistently shows heavier peaks in cancer DNA and its pieces. But where 17OH and 15NH2 are involved the aminations and hydroxylations consistently show smaller masses in the mass spectra of cancer DNA and its pieces. The larger massive pieces during methylations result and are explained by the addition of more massive 13CH3 into the functional of DNA nucleotides. The less massive pieces during aminations and hydroxylations are explained as resulting from loss of more massive 17O and 15N from the functionals of cancer DNA and its nucleotides. In general, the 13CH3 and its positive NMMs strengthen the covalent bonds in cancer DNA for binding 13CH3 is a stronger nucleophiles for more rapid replacements in DNA and its nucleotides. But the 15NH2 and 17OH and their negative NMMs weaken the covalent bonds in cancer DNA for bond breakages and 17OH and 15NH2 are better leaving groups for more frequency of 15N and 17O of nucleotides under electromagnetic fields during NMR analysis to explain these observed mass spectra.
It may not be that 17O and 13C attract or repel by internal C frame magnetism. It may be that they self conform to form quanta. So all + NMMs → classical or all – NMMs → classical, but balanced + NMMs → and – NMMs → quantum and the monopoles separate locally but bind globally. So on one scale they may bind and on larger scale repel or vice versa. So 14N drives biomolecules by imbalance perturb e- e- quanta 15N may disrupt such natural imbalance of 14N; 17O also disrupts the 14N imbalance; 13C disrupts e- e- quantum mechanics; and 14N cannot help 13C. But 17O can help 13C at higher temperatures, in electric fields and magnetic fields. But 15N can help 13C at higher temperature, in electric fields and magnetic fields. 17O disrupts 15N quantum mechanically, but together they help pull in 13C and less 14N causes loss of protein nuclear perturbation. On such basis the author notes tumors may be killed by enriching 17O, 15N, and/or 13C in their biomolecules and exposing them to strong electric fields and/or strong magnetic fields. 13C may overdrive classical mechanics of protein with 1H and 14N. 13C causes accelerated glycolysis as driven fragmentation of glucose. But the combining of C to O is opposed by 13C and 14N in the Kreb cycle or they oppose sp3→ sp, sp2. + NMMs favor sp3, - NMMs favor sp and sp2 for 13C but not for 17O. So 13C favor sp3 and 17O favor sp3 (for different reasons) as higher e- e- density for 13C increase electron density on C and less e- e- repulsions for negative NMMs of 17O reduces electron repulsions about O. So 13C and 17O accelerate glycolysis by one environment. But 13C and 17O suppress the Kreb cycle as in the Kreb cycle the sp2 and sp hybridizations are catalyzed about C and O and the 13C and 17O oppose such sp and sp2 hybridizations but favor sp3 hybridizations. But 17O and 13C decelerate Kreb cycle by different environments.
In this work, the author proposes a new way to alter functional groups of uridine, thimine, cytosine, adenine and guanine (by isotopic substitutions/replacements of !H, 16OH, 14NH3, 12CH3, and 24Mg by nonprimordials of 17OH, 15NH3, 13CH3, 2D and 25Mg) as nonprimordial, functional groups entering and to replace primordial, functional groups of nucleotides by this new theory as by the many aromatics of the purines and pyrimidines oscillating their electrons to couple the many nonzero NMMs of these nonprimordial, functional groups for activating their nucleophilic substitutions of primordial, functional groups. The theory 1, 2, 3 introduces novel chemical dynamics of multiple electrons and multiple functional groups in nano-domains behaving nonclassically to couple their spins and electronic motions to violate the 2nd Law of Thermodynamics momentarily as energy is focused into specific fewer atoms of the group to catalyze transportations, transformations and momentary transmutations for novel chemical dynamics of many bodies as the nanodomains by this theory gets quantum mechanically into a single atom or small molecule by Little Effect the fermionic atoms by their nuclei (NMMs) are in analog to fermionic electrons in atoms. By such the atoms in the domains have a wave natures and they exchange and correlate to move and alter their wave natures and they exchange and correlate to move and alter motions and positions in the nanosolution so as to lower energies. But for biomolecules such waves are quantum waves and differ from larger classical waves as by the theory 1, 2, 3, the nano, subnano waves can superposition to focus intensites in to specific bonds for quantum activations and this explains novel bond activations by enzymes. Such motions and altered positions manifest new chemical changes of the atoms, small functional groups in the nano-domains of proteins, nuclei acids and nanowater and nano-ammonia. So that the biochemical transformations have been previously described by the author as nanoscale quantum wave mechanics that manifest at lower temperatures for fermionic nuclei having nonzero NMM, but higher temperatures and pressures and E, B can induce the quantum wave mechanics of nanosolutions composed of null NMMs.
So inside the nucleus, GATC are the nucleotides; but outside nucleus GAUC are the nucleotides. Methylations (13CH3 ) of U cause T*. So isotopic effects in cytoplasma get into nuclei by U + 13CH3→ T* in cytoplasma and transfer of T* into nucleus. So 13CH3 on T* in nucleus causes altered genetics as reasoned by this theory. In prior work, it was previously published U expresses as T* due to 13CH3. So 13CH3 seems like H (by their positive NMMs); so T* becomes as U; and U in nucleus alters genes. Normally U is in cytoplasma and T is in the nucleus. So by U → U* → T*,U* is transport into the nucleus via T*, the replication of DNA is altered by such U* and T* in the nucleus of cells as 13CH3 (methyl) on the thymine alters biochemical dynamics. Also 13CH3 in T* may accelerate T*→ C* by dehydroxylations, deaminations, and aminations. So this causes mixing of nucleotides and mutations by chemically interconverting of nucleotides. T → U. U → C. Such chemical transformations of nucleotides alter the genetic code to cause cancer and other diseases. This theory 1, 2, 3 further proposes that the external static magnetic fields and radiofrequency fields can excite these nanosolutions to accelerate these nonprimordial substitutions. It may be that such chemical transformations of nucleotides in normal cells to mutate normal cells to cancerous cells are kinetically and thermodynamically possible by a few nonprimordial substitutions; but with more and more nonprimordial substitutions, the replacements are slower or not allowed. Such chemical transformations may occur as normal cells transmute to cancer cells with higher amounts of NH3 in the cancer environment. But this theory proposes that the use of external magnetic fields for stimulating cancer cells so their DNA pull in more nonprimordials so the excess nonprimordials kill the cancer. With such rapid replications of cancer DNA, it should be easy to disrupt the genes in cancer so the cancer cannot produce its proteins for glycolysis to kill the cancer.
Adenine is unique as it is the only nucleoside lacking O group and has only N functionals. The N is weaker base and weaker nucleophile than O as in guanine, uridine, thymidine and cytidine. It is on this basis of RBL that the 17O in water is the basis for the enrichment of 17O in DNA and RNA. The 17O in the many rings help the ring pull in 13C as by 17O activating bond cleavage of 17OH and + NMMs but many 14N, 1H and 33S and other 13C can induce, new bond formations, but as excess + NMMs cleave + ... + NMMs bonds and excess – NMMs cleaves - ... - NMMs bonds in quantum fields. So quantum fields + … - NMMs globally bond and + ... + NMMs locally agitate bonds and - ... - NMMs locally agitate bonding and as the nonprimordial isotopes clump they manifest new enzymatics of the DNA and RNA. . So this theory 1, 2, 3 introduces totally new chemical dynamics as here it is determined novel nonlocal chemical bonding but local chemical decomposition and/or nonlocal chemical decomposition but local chemical bonding.
The patterns of null, + and – NMMs (needles in haystack) can cause local bonding while globally the fermions are unbound. So the theory 1, 2, 3 determines that systems of + and – NMMs (Nuclear Frames) bind the atoms globally on large scales as they locally repel and are chemically broken. This is why 13C and 17O and 15N activate transition states and lower the barrier to chemical substitutions of isotopes. But the theory 1, 2, 3, the + and - NMMs as are more common in our sector of the Universe (or in other sectors – and - ) locally on nuclear scales repel but on global scales they bind/attract. So this also in other sectors of Universe with – NMMs have – NMMs interacting with – NMMs repel locally in nuclei but bind to attract globally as in Ag nanoparticles and other rare elements having all – NMMs. But such considerations, RBL gives a totally new model for transportations (superconductivity) and transformations {chemical and biological dynamics}. So prior chemistry and transport have focused primarily upon + ... + NMMs and the globally binding by e- e- and the locally repelling /unbinding by NS Frames with less chemistry and transport possibilities. Such manifest in primordial nanosolutions in cells having + NMMs of 14N, 1H and 31P and null nuclear magnetic moments (NMMs) of 12C, 14N, and 16O and normal primordial biology manifest on such basis of repulsions on NS Frames and motions and biochemistry of binding on L frames of wavefunctions. But RBL introduces totally new effects of – NMMs + ... + NMMs binding locally in NS Frames and repelling globally in L frames. So bonds are broken globally to isolate the e- e- but locally the e- e- bind by the + NMMs and – NMMs to manifest a Reggie Pair bond by NMMs of + and – NMMs as this occurs in nanosolutions in cells as 17OH2 and 15NH3 enrich with 13CH3 in the nanosolutions, proteins and nucleic acids. So the nanosolutions bind on NS Frames but globally the e- e- are more broken chemically. So the proteins and nucleic acids have different motions, binding enzymatics and biochemical reactivity. Such theory explains the cancer cell as the protein ··· nucleic acids interactions are altered by the + and – NMMs causing wavefunctions to repel. But the nuclei still pin the atoms together for cancer habitat.
It is important to consider that by such model of theory 1, 2, 3, in normal cells the 14N and 1H can modulate the bond cleavages and bond formations of PO3- and the ribose as the compressions may induce bond cleavages of 31PO3- to release energy and the chemical composition of ribose (of null NMMs). As compressions break + NMMs of PO3-, but bind C-C-O-H of ribose of O (null) NMMs. But then the rarefaction binds PO3- and fractional fissings and fusings decompose ribose and these can couple to pull apart base pairs or also such dynamics couple to surrounding proteins to bind or decompose the proteins to pull in or push out proteins. And such can explain DNA replications quantum mechanically as bases recognize quantum mechanically by patterns of NMMs and compress/rarefy with pulling in and pushing out. And likewise for transcriptions. And in ribosomes such act vice versa as pulling in amino acids under conditions whereby the oligonucleotides, RNAs are stable.
The clumping may help 15N incorporations into the oligonucleotides. The functional groups can dynamically shift the functionals to find equilibrium with the kernelling of nonprimordials, lowering the energy relative to random distributions of the nonprimordials in normal cells. Such clumpings of dense regions of nonprimordials isotopes alter nuclei acid bindings, bond strengths and chemical stabilities as by enzymatic actions on the kernel regions. But the clumps in normal cells may be linked to noncoding regions of DNA. So later the oligomers of food tannins can modify the functionals in cancer cells more than in normal cells to kill the cancer cells!
The guanosine may be more reactive due to 17O relative to 15N as the 17OH is better nucleophile than the 15NH3 and 15NH3 or 14NH3 is less abundant in normal cells! It could be that the presence of 14NH3 causes the genetic alterations of normal cells to cancer cells and the 15NH3 helps as by mutating genes. Comparing the various signals, the FWHM of signals from fragmented DNA in normal cells appear broader relative to the signals of fragmented DNA from cancer cells (note that this points to clustering of nonprimordials in cancer DNA and this narrow FWHM of cancer DNA is consistent with clustering of nonprimordials to dense kernels in the cancer DNA). The smaller FWHM in cancer DNA fragments may be near and from the clumping of nonprimordial functional groups of deuterons, hydroxyls, amines, and methyls. Such clumpings of nonprimordials lead to sharper distinct fragmentations during the mass analysis of DNAs for sharper peaks relative to broader peaks in fragmenting of the primordial regions of normal DNA. By the theory, the incorporation of nonprimordials of 2D, 13C, 15N, 17O and 25Mg into cancer DNA by functionalizations and defunctionalizations of the nucleotides appear to explain these observations of DNA isotopic differences between cancer and normal cells. It is important to note that the easier fragmenting of these pieces having nonprimordial isotopes in cancer cells relative to less sharp fragmenting in normal cells is evidence of altered interactions of nonprimordial isotopes in the DNA and RNA for altering the replications, transcriptions and translations.
So after considering these different causes of the functional groups in cancer and in normal cells on the basis of based on the spectra, a discussion of the proclivity of nucleotides and oligonucleotides to the new chemistry is next given. The aromatic and the ring structures by the theory 1, 2, 3 previously modelled such biomolecules on the basis of Na+ and K+ interactions with graphene oxides. It was determined that Na+ and K+ NMMs interact favorably with graphene oxides with their sp2 and sp3 mixed hybridizations and magnetics via the nonzero NMM of K+ and Na+. Thereby, likewise, RBL reasoned similar NMMs interact with sp2 and sp3 networks but now in biomolecules like DNA. So that the theory 1, 2, 3 introduced changes in interactions in the DNA as primordials of 1H, 12C, 14N, 16O, 24Mg, and 32S are replaced by nonprimordials of 2H, 13C, 15N, 17O, 25Mg, and/or 33S of different NMMs. Such manifest as the purines and pyrimidines in nucleic acids regions with sp2 aromatic and regions with sp3 nonaromatic in analog to prior different regions in graphene oxide.
Why Do Nucleotides Transform on Atomic Scale
Thereby the theory 1, 2, 3 realized nuclear spins could couple to carbon covalent dynamics in prior graphene and in biomolecules. But even before the experiment with graphene oxide the spin interactions and NMMs of p+ interacting with biomolecules had been published in a book Chapter 2. So by considering graphene an analog for proteins and other biomolecules. The theory 1, 2, 3 proved that nuclear spins in general can couple to biomolecules to alter catalysis and enzymatics of biochemical reactions. Next in this work, the mechanism by Little’s Effect are given for driving the replacements and substitutions of null NMMs by nonzero NMMs. The more extended aromatic rings may couple spins of the nuclei for faster clumped, accelerated isotopic enrichments of the ring systems via the aromatic π electrons as the aromatic electrons couple the separated nuclear magnetic moments (NMMs) and induce transports, exchanges and replacements of the different NMMs. These extended π electrons and orbital exchange and bonding about many atoms may be mechanism for more strongly coupling the nuclear spins and NMMs {Reggie Acids and Bases of electron radicals (fermions) and nuclear spins (fermions) and nuclear radicals and orbitals} to orbitals (of Lewis Acids and Bases, both electronic and nucleonic) via the exchange by π electrons. The nuclear spins and the nuclear orbital angular momenta are thereby exchanged and coupled via delocalized π e- e- in the phenyls, polyphenols, and polyphenylamines. Also by this model 1, 2, 3, such spins are not limited to e- spins; nuclear spins are also coupled, transformed, transported and transmuted by π e- e- and d orbitals of transition metals. By the model 1, 2, 3, the substrates couple and quantum mechanically exchange the NMMs in the enzymes and macromolecules and vice versa.
The localized bosons, the localized fermions, the delocalized bosons and delocalized fermions may be driven by surrounding thermal perturbations, gravity, electric, magnetic and QF driving forces. The relative stabilities and interactions for stable ferromagnetism, paramagnetism and diamagnetism are by Little’s Rules as diamagnetism in such systems may obey Little’s Rules 1 and 3 but ferromagnetism, antiferromagnetism and paramagnetism in such systems may obey Little’s Rules 1 and 2. The diamagnetism may be by the bosons localized as in diamond, but in graphene the bosons are delocalized bosons. Such happens in graphene to cause electronic spin paired fermions in the delocalized electrons. These unpaired delocalized fermions cause the delocalized to rehybridized to localized as sp2 to sp3. The theory of RBL determines some transient spin induced, finer, azimuthal, fractional, continua quanta numbers for transition stages during transportations and transmutations. And likewise with the nuclei, as the nuclei interact with the electrons and bosons in graphene the nuclear spins and orbitals angular momenta in nuclei alter the electronic delocalization for singlet to triplet on other spins. The fractional fissings and fusings of nuclei seep QF into electronic shells as by the theory 1, 2, 3, so as to transiently create ultrafine continua of azimuthals for mixing, coupling, transporting, transforming and transmuting electrons for novel superconduction, chemistry and catalysis/enzymatics. Vice versa e- e- rehybridizations and spin polarizations can alter the couple nuclear orbital momenta by RBL Effect. The localize bosons verses delocalized bosons allow different coupling of nuclei and their NMMs. The thermodynamics may favor one or the other, but the change from one to other involves kinetics and dynamics by Little’s Effect. The e- spins and nuclear spins via delocalized or d (azimuthal) π e- e- can couple to alter the symmetries and motions from locals to nonlocals and vice versa.
Why Purines, Pyrimidines, Polyphenols and Polyphenylamines More Strongly Couple NMMs?
Pure metal clusters and nanoparticles may also couple nuclear spins. But in molecular compounds, the coupling may not be possible via more localized molecular orbitals. But the delocalized molecular orbitals via π bonds may afford the delocalized bonding over many C, N, and O bonds as previously proposed in theory (RBL ferrochemistry). So that the π electrons can couple spins and orbitals of electrons (e- e- Lewis pairs and radicals) and the π electrons can also couple the nuclear spins and nuclear angular momenta over many atoms in nanodomains. Thereby the pyrimidine’s aromaticity more exchange the nonprimordials. The purine’s aromaticity less exchanges the nonprimoridals. Just as for the pi electrons in purines and pyrimidines delocalize the NMMs of 13C, 15N, 17O, likewise the pi electrons in polyphenols and polyphenylamines delocalize these nonprimordial isotopes and their NMMs. The nucleotides, oligonucleotides and nucleic acids couple their nuclear magnetic moments (NMMs) with NMMs in surrounding nanowater and accumulated NH3 to accelerate primordials replacements by nonprimordials by different NMMs. So the delocalized e- pull in NMMs. These molecular orbitals can couple spins on centers. So also spins can alter orbitals and the orbitals can alter spin centers, spin ... spin orbital interactions not only alter orbits but flicker spins; transition states break bonds; spins flip and intervening metal orbitals and/or orbits couple spins to other regions when orbits change and spin pairs change polarizations to change bonds. By this mechanism 1, 2, 3, the spins not only interact with the orbits, but the spins transform by fractional, reversible fissing and fusing. Fissed spins fractionally, reversibly fiss and fuse to orbits and vice versa the orbits fuse to spins. So also NMMs via e- e- orbitals can couple nuclear spins and change the orbitals. Nuclear spin momenta and orbital momenta can alter the e- e- orbital. And e- e- orbitals can alter nuclear angular momenta. RBL here notes NMMs are variable by not only during chemical reactions but also during chemical reactions, enzymatics, vibrations, optics and e- e- transportations and transmutations. The nuclei are perturbed so relative motions of nucleons change and the nuclei swell and compress for fractional, reversible fissing and fusing to alter and to couple to surrounding e- e- lattice. Thereby momentary changes in NMMs occur. The DNA and RNA have more pi bonds for easier activations by breaking pi bonds for easier replacing isotopes relative to other biomolecules. Therefore, it is this reason of the aromatic rings in purines and pyrimindines that the nucleotides in DNA more readily exchange isotopes nuclear spins and NMMs relative to other biomolecules.
It may be possible by such unique ability of DNA and RNA via their nanodomains of graphene, diamond, alkyl, aromatic and/or diamagnetic, paramagnetic ferromagnetic functional substances that the resulting DNA and RNA catalyze isotopic exchange in proteins. So that during DNA, RNA and protein bindings, interactions, charge exchanges and enzymatics, isotopes may be exchanged. By this theory, the 14N, 1H and 31P via fractional, reversible fissing and fusing cause the denatured proteins to renature and the DNA to unnature and renature during reproduction, and RNA to denature and nature. So in general, the NMMs in the proteins and nucleic acids cause orbitals to change. So the proteins and nucleic acids denature and renature. So the proteins and nucleic acids renature so rapidly due to huge fields caused by the nonzero NMMs of 14N and 31P within them (and 1H2O in surrounding nanowater). Thereby from this theory the RNA may catalyze the nonprimordial replacements in amino acids as the RNA translates proteins.
The isotopic exchange is selective in uphill anabolism in animals and humans nonadiabatically as it is selective in uphill anabolism in plants adiabatically. It is during uphill processes of DNA replications, RNA transcriptions and protein translations that the proteins are isotopically altered. Virus RNA can modify so the RNA produces unhealthy proteins. It is that the side chain sugar and side chain phosphate couple energy into the nucleoside to break bonds. It is that the side phosphates and side sugars help the NMM replacements. This occurs by the ferrochemistry of the bond rearrangements of the sugar releasing energy reversibly as accumulations and absorbing into the phosphates by NMMs and other oligonucleotides by 14N and 1H so as to give energy to promote the dynamics. So activated states near or far are involved and then as the transition states relax to products, the phosphates collect the energy and restore it back to the sugar unit. There is chemical energy in the sugar and the phosphate can store chemical energy and the nucleosides can delocalize energy. It is on this basis that some viruses can kill cancer cells. But the downhill catabolism (relative to uphill anabolism) is less affected by isotopic replacements as the electronic energy can drive and dictate the dynamics. But in glycolysis the down hill is accelerated by the isotopic replacements as downhill glycolysis is reverse of photosynthesis in plants so the downhill accelerated by nonprimordial 13C just as uphill is slowed by 13C. It is logical that exothermic downhill is less discriminating nonprimordial / primordial replacements. But in Kreb cycle, higher electric and magnetic fields in the substrates and the enzymes cause stronger effects on the downhill processes as the high fields can couple more strongly to the NMMs for the nonadiabatic Kreb cycle so that Kreb cycle becomes adiabatic as the heat is organized in the high fields. This is the reason the Kreb cycle is more sensitive to nonprimordial isotopes relative to the glycolysis process.
Thereby this theory determines that the DNA may accumulate the nonprimordials from the proteins and sugars combusting and then the DNA may incorporate the nonprimordial isotopes into the proteins during translations, replications and transcriptions for the nonessential proteins. The eating of nonprimordials in nonessential proteins can cause the animals to accumulate nonprimordials; first in nucleic acids and then in proteins via nonessential proteins. But as the organisms eat other animals and obtain essential amino acids, then the essential amino acids have more nonprimordials. So the nonprimordials within the eaten essential amino acids connect to alter catabolism in cancerous ways in the essential proteins. The 13C in lysine is crucial for animals and humans to develop cancer. So diet accumulate 13C in DNA and then diet of essential amino acids accumulate nonprimordials in enzymes. When the two conditions optimize then cells become cancerous. Cancer cells may accumulate nonprimordial isotopes until they die and then the innards with the nonprimordials of the dead cancer cells are eaten by normal cells and the the surrounding normal cells transform to cancer cells. This may be a basis for metathesis. RNA with the nonprimordial isotopes can synthesize nonprimordial amino acids and construct nonprimordial proteins. These with lysine can cause cancer.
More general discussion is given here of NMMs coupling by MOs and AOs causing nucleophile substitutions and NMMs undergo substitutions and replacements. Nucleophiles driven by nucleophiles but the spin driven by magnetism as the null spins diamagnetically pushed out MO and the nonzero spins pull in or push out MOs and AOs. But what about the + NMMs and – NMMs. The + NMMs pull in + NMMs and push out – NMMs in MOs and AOs. But in nuclei and continua + NMMs push out + NMMs and pull in – NMMs. So thereby 13C is pulled into other 13C via π bonds as the many 13C nuclei create self conforming MOs. But 17O disrupts MOs of 13C to activate bond rearrangements. As 17O pulls 13C nuclei together and yet push their QFs apart for driving bond activations for bond rearrangements. This is powerful as by this theory 1, 2, 3 introduces new types of interactions as two or more objects interact in counter ways on different states on L frames they attract but smaller frames they repel and/or on L frames they repel and on smaller RS frames they attract. Something on inside binds whole and whole repels. Or something on inside repels as whole binds! This is new by author for how particle ⟷ wave. This is new basis for compositie forces. The author published this in 2007 as p+ and nuclei bond e- e- of covalence by fissing of p+ and nuclei to create QFs to bind the e- e- pairs. So it is that the 17O can attack as it breaks up many + … + ... + ... + ... + NMMs. This may explain Ag nanoparticle atomizes due to the interactions of all its negative NMMs. So now this 33S, 2D, and it 14N help pull in 13C and 17O as such lowers Eactfor such isotopic replacements inside organisms for replacements of primordial isotopes by nonprimordials. But what about 15N; it lowers Eact at higher temp but at lower temp it pushes 13C away.
The observed higher deuterations, methylations, aminations, hydroxylations and enrichments with 13CH315NH2 and 17OH in cancer cells is consistent with nonrandom clustering and higher density methylations in DNA of cancer cells 10. Moreover, in this work in addition to explaining faster methylations by 13CH3 to cause cancer, the faster methylations are explained in details by the accelerated 17OH hydroxylations and many body 15NH2 aminations, causing the transformations of C →T → U and A → G for chemically altering DNA and RNA for new chemical paths of DNA and RNA mutations for explaining cancer. 13CH3 may be causing more methylations of DNA in cancer cells as in this theory 1, 2, 3, 13CH3 is a stronger nucleophile than 12CH3. This isotopic effects of the nucleic acids can explain recent mysteries. Positive NMMs of 13CH3 relative to null (0) NMMs of 12CH3 by the theory 1, 2, 3 cause the e- e- to be pulled closer to nucleus of 13CH3 relative to 12CH3 nuclei. 24Mg2+ should interact less strongly with 13CH3 (and make 13CH3 a stronger nucleophile) relative to 25Mg2+ for more altering the bonding of 13CH3 by 25Mg2+ in cancer cells relative to the weaker effect of 25Mg2+ and/or 24Mg2+ interacting with 12CH3 in the normal cells to explain the observed selective killing of cancer cells by 25Mg2+11. This accumulated 13CH3 in RNA and DNA then alters RNA and the RNA alters translated proteins for mechanism of splicing phenomena. 12. This theory of RBL determines the chemically altered RNA by nonprimordial isotopes causes the splicing of proteins that is hallmark for cancer genesis and habitats. The transmuting of 12CH3 to 13CH3 of the space twin relative to the earth bound twin would explain the observed elongation of the telomeres of the space twin as by 13CH3 methylations of his telomeres and stronger binding of his telomeres by 25Mg2+ for elongation rather than shortening of the telomeres of space orbiting twin 13. The stronger binding of the telomeres containing 13CH3 may less frazzle the ends for continued elongations.
Methylation and Altered Binding and Transcriptions and Translations
So after reasoning and explaining how the nucleotides are isotopically mutated and some consequences, here it is considered how altered genes malfunction. So these alterations of nucleotides alter the sequencings, constitutions, connectivities and stereochemistry of isomers so what are consequences? Based on this model 1, 2, 3, the methylations of the cytosine not only causes the cytosine not to bind guanosine, but moreover the methyl-cytosine may be misread as thymine and vice versa the thymine may be misread as methyl-cytosine. These are some of the consequences of changing the isotopes in nucleotides. The normal base pairs are GC and AT pairs. Also the functionalizations / defunctionalizations can alter the DNA and RNA sequencing transformation C → T→ U and A →G so as to alter DNA and RNA and alter proteins for changing RNA, DNA and proteins content in cells to damage cells. So C may be methylated similar to A and OH- may replace NH2- for C → U. So methylation of C and deaminatation forms C and U for mutations and for consequent possible misreading of protein; so for example UUC (Phe) → UUU (Phe), CUU (Leu), CUC (Leu), CUA(Leu), and/or CUG (Leu) → UUU (Phe), UUC (Phe), UUA, UUG (Leu). So in some cases U and C can interchange without misreading protein, but in other cases such changes cause misreading and mutations. Likewise mutations as C → T and → G → A can cause splicing of proteins as by the change in translations of amino acids. Thereby chemically interchanges in nucleoside sequences change the selection of peptides to alter proteins. Stops in nucleic acids do not involve C: UAA, UAG, UGA! The creation of organisms may have intentional avoided C in stops as the mutations of C would affect stops. The G is in stops and mutations of G may cause stops in nucleic acids not to stop for a basis or cancer. Thereby DNA is altered by nonprimoridals substituting for primordials.
The 13CH3 binding may alter interactions and dynamics due to its different NMMs. Although thymine already has a 12CH3, by changing the 12CH3 to 13CH3, the properties of the thymine change so that the 13CH3 may appear like H on the phenyl ring of thymine; so thymine appears to behave like uracil for altered replications, transcriptions and translations. Such 13CH3 and its + NMM may appear as 1H so the thymine in nucleus of cells appears like uracyl (U) with alterations of the DNA replications and transcriptions inside the nucleus. So it is that U can exist in the nucleus but thymine (T) exists only in the nucleus. But if 13CH3 replaces 12CH3 on thymine then thymine appearing as uranyl can exist in cytoplasma also to alter biochemistry in cytoplasma. And uranyl can methylate to enter nucleus. This means that uracyl in the nucleus can be template as thymine (as 13CH3 in thymine causes it to appear as uracyl). Or the other possibility is that the 13CH3 in thymine causes it to not be recognized. A third possibility is that the 13CH3 causes the similar nuclear behavior as H so the thymine may behave as cytosine assuming the =O (OH) and NH2 manifest similar basic interactions. So these are possible consequences of 13CH3 on the thymine.
C and G Depletions and Cancer Defient Amino Acids in Cancer Cells
In addition to these nonprimordial, induced misreads of nucleic acids and proteins and nonprimordial, induced, inter-chemical transformations of C →T → U and G → A, this mechanism 1, 2, 3 further determines the consequent deficiency in C and G due to the + NMMs of 13CH3 and – NMMs of 17O and 15N by difficult hydroxylations of A → G due to 17O and difficult demethylations of T → C. The consequent dynamics cause deficiencies in C and G 14, 15, 16, 17, 18 in cancer cells. The deficient C and G 14, 15, 16, 17, 18 on basis of this theory 1, 2, 3 causes deficient proteins translated by C and G 14, 15, 16, 17, 18. See Figure 4. For instance, G and C strongly translate Gly (GGU, GGC, GGA, GGG) 21. {Note Gly and Pro are extremely important for alpha helical breakers. Gly and Pro start secondary structures of beta turns. Beta turns are turns in primary structure. Pro 19 has odd, cyclic structures in peptide bonds and these cause bendings of peptides. Gly has small size and can have large conformational changes due to lack of steric hindrance by Gly due to its small size. Bending breaks alpha helicies.}
Figure 4.Nucleotide Codons for Amino Acids (reference 9)
The Arg 23, Try 22 and Ala 20 also have strong translations by G and C and shortages of C and G in cancer cells are here reasoned to cause cancer habitat and transform normal cells to cancer cells: Arg* (CGU, CGC, CGA, CGG); Trp* (UGA, UGG), Pro (CCU, CCC, CCA, CCG); and Ala (GCU, GCC, GCA, GCG). 19, 20 Quite interesting, Arg and Trp are also essential amino acids; and this enforces this theory of the cancer genesis due to shortages of G and C and the inability to synthesize the Arg and Trp translated by G and C. But then other amino acids are marginally affected by deficient G and C: Leu (CUU, CUC, CUA, CUG); Val* (GUU, GUC, GUA, GUG); Ser (UCU, UCC, UCA, UCG); Thr (ACU, ACC, ACA, ACG); Asp (GAU, GAC); Glu (GAA, GAG); Cys (UGU, UGG); His (CAU, CAC); and Gln (CAA, CAG). * means the amino acids are essential amino acids. But then, the following amino acids are not strongly affected by shortages in G and C contents in cancer cells: Phen* (UUU, UCC); Ile* (AUU, AUC, AUA); Met* (AUG); Try (UAU, UAC); Asn (AAU, AAC); and Lys* (AAA, AAG). So on basis of such deficient G and C causing deficient templating of amino acids to form proteins the following consequences are reasoned. The predicted deficient Gly, Arg*, Trp*, Ala, and Pro correlates with recent analyses of microenvironment of tumors. Van der Heiden 24 recently observed microenvironments of tumors are depleted in Trp*, Arg* and Cys. It is also important to note that Try* and Arg* are essential amino acids, as cells cannot synthesize these essential amino acids. But Gly and Glutamate where found by van der Heiden 18 to be abundant in cancer microenvironments. But Gly can be synthesized. And Glutamate is nonessential and can be synthesized so this observed abundance of Gly and Glu are consistent with this theory of cancer 1, 2, 3. The stops are encoded by UAA and UAG, therefore excess A and may cause high densities of stops. The telomerase has its own RNA (3’ – CCCAAUCCC 5’) for translating teloemerase. So telomeres cannot elongate and this is habitual of cancer cells as 13CH3 methylates C on telomerase, then the C of telomere cannot help elongate the telomerase. It is important that the telomerase translation involves a lot of C and by this theory the deficient C may affect telomerase formation, length and stability for causing cancer as the lack of C causes lack of telomerase and the lack of elongating telomeres which is one hallmark of cancer.
After discovery of this new DNA, RNA and protein chemistry by Little Effect via NMMs of nonprimordials, this work considers plants oligomers and possibly such chemical interactions of plant oligomers with human oligomers. This work determined that just as the RNA, DNA and proteins can undergo intrinsic internal accelerated methylations, deaminations, aminations, hydroxylations and deuterations of nonprimordials relative to primordials; then also foods having similar oligomeric structures can also exchange primordials and nonprimordials via functionalizations and defunctionalizations between dietary oligomers and nucleotides in the host. But what happens to DNA as animal products are consumed? Plant products have less nonprimordial, 13C, 15N, 17O and 2D in their nucleus acids. But how does such low nonprimimordials compare to animal DNA? Animals tend to have in general greater amounts of 13C and 15N relative to plants. Scientists find link between plant telomere and human telomere so plants live longer as by their lower nonprimordials relative to animals and humans. The diet and metabolism of tree differs from animals and trees have less motion and less energy demands so trees do not break down 13C compounds and then construct 13C as much in their DNA for high nonprimordial contents as occurs in animals and humans; so tree DNA less mutates so trees live longer. The theory here determines that penalty of motion as by needed catabolic metabolism is breaking nonprimordial molecules and consequent uptake nonadiabatically of nonprimordials into DNA with mutations. Muscles and lysine cause 13C and cancer. Trees and plants use sunlight and operate adiabatically so less nonprimordials are taken up. Heat may help animals and plants pull in nonprimordials, plants operate cooler and pull in less nonprimordials. This explains how animals mutate DNA and develop cancer. This leads to cancer in humans and animals.
In this theory RBL tried to correlate cancer to motion and diet on this basis of nonadiabatic catabolism and uptake of nonprimordials by animals and humans. So eating cancerous DNA may also cause cancer to be transferred to host DNA and RNA. So cancer can be transferred by large transfer of cancerous tissues. Rats are implanted with cancer tissue with induction of cancer. In this work, it is reasoned that cancer cells of different types may kill each other. Injecting different types of cancer into a tumor may kill the tumor as the DNA and RNA of the two cancers differ. It may be possible to kill tumors and cut it out by surgery. The nonprimordials are determined to accelerate such new chemistry by differences in kinetic and thermodynamics of functionalizations and defunctionalizations. Thereby a new chemistry is described based on discovered for nucleotides based on NMMs and magnetics driving substitutions of NMMs and a mechanism 1, 2, 3 by which NMM substitutions can couple and mix with nucleophilic substitution energies. So by replacing new oligomers with nonzero NMMs in heavier isotope, the DNA of normal cells is disrupted to cause cancer. But in this work, it is further determined that just as the isotopic accumulations can transform normal cells to cancer cells, excessive nonprimordials can accumulate to kill cancer cells. Grape seeds may kill cancer but they may cause cancer as in this work, the grape seeds have oligomers of proanthocyanidins, which are in this work determined to have excess 13CH3, 17OH, 15NH2, and/ or 2D that can replace 1H, 12CH3, 14NH2, and/or 16OH in cancer to oversaturate the cancer DNA with nonprimordials to kill the cancer.
It has already been published in 2007 1 that the + NMMs and nuclei of 14N, 16O and 12C via the proton (1H) can magnetically couple for novel many-body nuclear magnetic moments (NMMs) and nuclear orbitals to cyclically move, transform and transmute for normal anabolism and catabolism. Thereby it is determined that without such effects of the protonic nuclei, life cannot exist and thereby disease may be caused by altering this natural rhythm 1. For instance, consider intrinsically the 31PO3 gives P center strong ability to attack ADP and AMP. So if change 12C to 13C then 13C makes 13CH3 a stronger nucleophile; and if change 16OH to 17OH then 17OH is a weaker nucleophile and if change 14NH2 to 15NH2 then 15NH2 a weaker nucleophile. So just as there is intrinsic NMM chemistry of 31PO3 then there is new chemistry by NMMs in 13C, 15N and 17O; 13C attacks more than 12C; 17OH attacks less than 16O. 17OH attacks differently than 16OH attacks; so it is not that 17O does not attack, but 17O attacks differently than 16OH. 13CH3 attacks more with 17OH than with 16OH. 17OH attacks 15NH more. It is that + NMMs attack + NMMs more in L Frame but less in L Continua and Nuclear Frames (NS Frames). {Where interior quarks are in QS Frames; quarks are inside hadrons in RS Frames; hadrons are inside nuclei in NS Frames.} The NS frames couple continuously to interior LS continua of the electronic lattice outside the nuclei. Electronic orbitals exist in L frames (discontinua); Electrons manifest continua about them for ES Frames and discontinua within the electron for E Frames. The ES frames of the electron can couple to the outer L continua of the electronic lattices to mix with the inner L continua of denser NS frame fields and such mixing of ES Frames and NF frames with diminution of stretch; transform and combine with other outer L Continua of other atoms, leptons and hadrons to manifest the C Frame (macroscopic frame) of magnetic fields, gravity, electric and thermal fields and spaces}. It is that + NMMs attack – NMMs less in L Frames and more in NS Frames. So thereby pressure effects manifest as high pressures push then the + NMMs into – NMMs; so the L Frames – NMMs repel + NMMs → NS Frames + NMMs attracting – NMMs at higher pressures.
This is consistent with RBL theory of high temperature superconductivity and why high pressures cause superconductivity. So now also with cancer as the cancer involves changing pressure the cancer may not metabolize as well; and by theory 1, 2, 3 this explains the changes in cancer as the host moves from surface of earth to outer space to kill the cancer due to changes in gravity and pressure. By this theory, the primordial isotopes of 1H, 12C, 14N, 16O, 24Mg and 32S manifest in normal cells at earth surface and atmospheric pressure with all positive NMMs; so all positive NMMs attract in L frames. But as cancer forms by 2D, 13C, 15N, 17O, 25Mg and 33 S on the earth’s surface, then the nuclei have + and – NMMs and + and – NMM repel in L frames. But if the normal cells and cancer cells are accelerated into outer space then the lower pressure as gravity becomes zero and the lost gravity in outer space less pushes + … + NMMs of normal cells to NS Frames for repulsion, so normal cells are less affected by earth’s gravity (this can also be a basis for new magnetic sensing of earth’s magnetic field by normal cells.) So now cancer cells on the other hand, have + and – NMMs and the loss pressure increases there + and – repulsions in L Frames to alter biochemical dynamics more in cancer relative to normal cells and may cause the cancer tissue to bind on larger scales for possibly killing the cancer. But such altered L Frames alter the glycolysis to kill cancer due to zero gravity. It is that 17O helps 13C in C Frame magnetically, but then 17O pushes 13C away in L Frames’ QFs. But then under compression 17O pulls 13C to it in inner L Continua and in NS Frames. This is how the 13C and 17O interact differently in complex ways on different scales to cause accelerated mutual replacements and substitutions for 16O and 12C in living organisms.
So the interactions are contravariant on different scales as attract on nuclear scale (NS Frame) and repel on QF scale (L Frames) and attract on magnetic frame of C Frame. It is that as something pulls another to it, but simultaneously can push it away simultaneously. Thereby this is new dynamics that by the theory 1, 2, 3 explains transformations of it and transmutations of it for new mechanics as introduced here by RBL. And this is how transport goes to transform and to transmute and vice versa. As transport is by push but if push so hard then it pulls as it pushes to stretch and pull it towards to transform it and to transmute it as by this theory of RBL! So 17O transmutes 13C where as 14N NMM pushes 13C away. 17O pulls 13C to its nucleus and pulls 13C atomic orbitals apart and magnetically binds 13C as it stretches its orbitals! This is a new type physics of chemistry as by nuclear magnetic moments (NMMs) and nuclear spins under very high temperatures, strong electric fields and strong magnetic fields 1, 2, 3. Simultaneous nuclear, chemical, and physical transformational phenomena are determined by the theory 1, 2, 3 to occur! So it is as the 17O internally binds it; it also stretches it and holds it globally! But 14N internally pushes 13C away as it binds it in QF and magnetically globally repels it!
So after considering/discussing the peaks and the nucleosides and how cancer DNA therefore has different peaks relative to DNA of normal cells and the different peaks are due to nonprimordials, next oligomeric in grape seeds 25 and the seeds are considered and compared to these nucleotides in normal and cancer cells. The novel chemical alterations of DNA and RNA by grape seeds oligomers are considered. The grape seeds are the cellular nucleic of the fruits with reproductive ability. So the biochemistry and biomolecules of grape seeds reproduction couple to biomolecules of human reproductions and malignant reproductions as by cancer so thereby the grape seed may couple to cell to cause cancer and/or to kill the cancer. The plants are observed to accumulate 2D, 13C, 15N and 17O in their proanthocyanidins in grapes with water deficit having more 17O and the 17O pulls in more 13C into seed 26. This is in the literature 26. It has been observed that the draught and 13C and 17O in the seeds make the seeds more anti-cancerous 25. What is it about grapes that they incorporate 13C and 17O among the plant kingdom? In this theory, it is determined that the chemistry of 13C, 18O and 17O cause greater incorporation of nonprimoridals in grape seed proanthocyanidins as by aromatic background network of the oligomers. The corn may have similar background oligomers to help it pull in more 13C in C4 process relative to C3 process 27.
Discuss Why the Grape Seeds Affect Cancer
In consistency of this reasoned aromatic, alkyl background network accelerating nonprimordial uptake by coupling NMMs as by this theory, researchers recently report larger plant oligomers have greater anticancer effects 7, 8, 15. The larger proanthocyanidins are more anticancerous as they have more nonprimordials isotopes and they pull in more nonprimordials; release more nonprimordials by extended C-C bonds and π bonds and/or bind nonprimordial isotopes existing in DNA, RNA and proteins and causing cancer and other diseases. Also consider that the enzymes of Kreb cycle may be able to pull in more nonprimordials relative to enzymes of glycolysis due to the high field substrates of Kreb cycle. Both such networks of changing covalence in Kreb cycle and changing covalence of glycolysis process and the oligomerics of proanthocyanidins manifest changing covalence in extended arrays of sp2 and sp3 covalence with intrinsic magnetics of the changing covalence and with embedded p+ and NMMs of other nuclei. By such Ferrochemistry the nuclei revolve to orbitals as by fractionally fissing their NMMs so as to couple the covalence and to alter the many covalence for breaking covalence by the many NMMs and pulling in nuclei and pushing out nuclei and rebonding covalence to new nuclei. Such explains the isotopic replacements by the covalent lattices with embedded NMMs in accelerated many-bodies relative to null NMMs as by the theory1, 2, 3. It is noticed that the greater enrichments of nonprimordial isotopes in the heavier isotopes correlate to the anti-cancer.
On this basis, a new idea 1, 2, 3 is presented. It may be that the different ferrochemistry of glycolysis, Kreb, replication, transcription, and translation can be reasoned by functional groups of amino acids as the alkyl + phenyl functionals may in proteins push together to induce greater nonprimordial uptakes. Also the 17OH and 15NH regions of functionals in proteins more push together to lower Eact for such 13C substitutions or to accelerate incorporations of 17O and 15N. So if the theory 1, 2, 3 looks at enzymes of glycolysis, it may find fewer Leu and Trp than in Kreb cycle. Kreb cycle may have more Trp and Leu so it would more incorporate 13C relative to glycolysis.
Proanthocyanidins are observed in grape seeds, cranberries and other fruits and vegetables 13. The greater amounts of proanthocyanidins (PACs) in grape seeds and cranberries are revealed in mass spectrometer as isotopic clusters are observed in Figure 5. The nature of the interflavan bonds (D2 amu) [M+Na}+ represented by observed masses. The PACs from grape seeds contain B type (m/z 1465) bonds. Masses represent variations in the nature of interflavan bonds (D2 amu) M + Na+. It is noticed that from the mass analyses, that the grape seeds explicitly show huge enrichments of either 2D, 13C, 15N and/or 17O in the mass spectra, but the authors of these prior data 13 do not correlate such properties of isotopes in the proanthocyanidins (PACs) to anti-cancer. Isotopes of predicted compounds are observed in the spectra with characteristic masses (m/z). For instance, the predicted monoisotopes for PAC of 5 DP with 4 B type interflavan bands are observed at 1465 m/z, which are observed to have primordials of 12C, 14N and 1H. See Figure 6- Figure 7. But mass of 1466 m/z is observed of similar intensity as the 1465 m/z for similar relative concentrations so that the 1466 m/z has possible contributions from possibly one 13C, one 2H or one 17O. The mass at 1467 m/z may have two of these nonprimordials 2D, two 13C or two 17O. The similar intensities of 1465, 1466 and 1467 m/z determine similar relative abundances and thereby isotopic enrichment of nonprimordials in the PACs. But in this work, the anticancer activity of proanthocyanidins is correlated with there enrichments with nonprimordial isotopes of 2H, 13C, 15N, 17O. Furthermore, the proanthocyanidins may be anti-cancer as by the similar chemical structures of the tannins and polyphenols to the nucleosides and the possible exchange of the nonprimordial isotopes between the nucleosides of RNA and DNA; and possibly more favorable binding of the nonprimordial enriched proanthocyanidins with nonprimordial enriched DNA and RNA in cancer cells. The proanthocyanidins may also alter the translations of proteins in cytoplasma and the synthesis of DNA in nucleus. In this work, it is determined that the nonprimordial isotopes couple (bind) more strongly to the cancer DNA and RNA relative to the RNA and DNA of normal cells, because by this theory 1, 2, 3 and data, the cancer DNA and RNA are isotopically different from the normal cells RNA and DNA. The stronger binding of tannin to cancer DNA is due to similar clumping of nonprimordials. The nonprimordials in grape oligomers may also chemically alter the DNA in cancer so as to alter cancer’s replication. Thereby the grape seeds provide the epigenetics to alter cancer DNA selectively so the seeds are anti-cancerous and this is the first molecular basis for anticancer properties of grape seeds. This is consistent with prior theory 1, 2, 3 for also treating cancer by the prior theory 1, 2, 3 as by the prior theory, it was proposed to use of nonprimordial enriched foods to selectively target the cancer. So the prior theory 1, 2, 3 looks at the DNA in the human and the cancer and finds the nonprimordials, and the prior researchers find nonprimordials in the seeds of grapes. So in this work, the nonprimordials in cancer and in grapes are correlated for anticancer activity of grape seeds. And the grape nonprimordials disrupt the cancer nonprimordials.
Figure 5.Oligomeric Proanthocyanidins from Adzuki Beans With Those Larger than Tetramers Showing AntiCancer Activities (reference 8)
Figure 6.Grape Seed Proanthocyanidins (PAC) Isotopic Reveal Isotopic Enrichments (reference 14)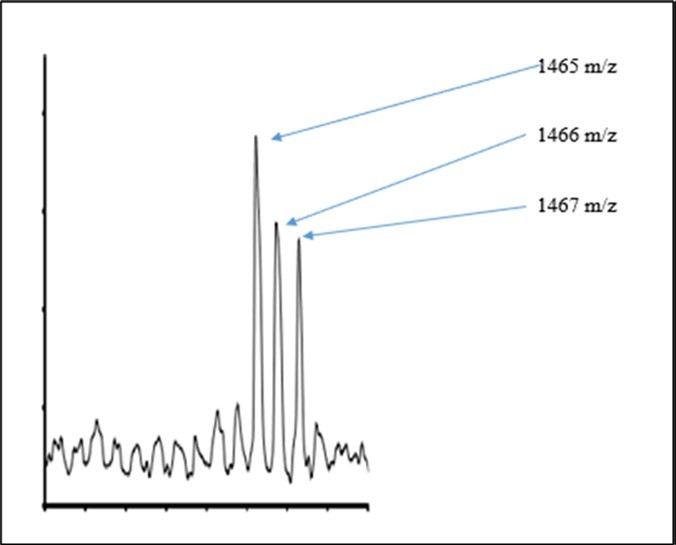
Figure 7.Structures and Masses of Nonprimordial Isotopes in Plant Proanthocyanindins (Reference 7)
Discussion of Possible Cancer Cure Relative to DNA, RNA, Protein and Proanthocyanidins
Previously the theory 1, 2, 3 proposed a treatment and possible cure for cancer by the patient eating normal food, but with added nonprimordial isotopes. Eating grape seeds is an application of this prior proposal by the theory 1, 2, 3 as grape seeds are in this work determined enriched in nonprimordial isotope. Therefore, eating seeds of grapes and other foods enriched with anthoprocyanidins are an examples of such eating foods isotopically enriched with nonprimordials. The theory 1, 2, 3 further proposed that the patients’ tumor should be irradiated with radio frequency of specific wavelengths so as to tune into the nonprimordial isotopes within the cancer cells with few effects on the normal isotopes and normal cells. The theory 1, 2, 3 proposed that radiofrequency rotates the nuclei and fractionally fiss to alter wavefunctions about to alter enzymatic activity within the glycolysis process to overheat and/or starve the cancer cells with few effects on normal cells. The theory 1, 2, 3 proposed that the patient’s tumor irradiated with specific X-rays of wavelengths tuned to excite in the near edge only nonprimordial isotopes for further inducing superluminous rotations of their nuclei to alter fields and quantum fields about to demagnetize and deactivate enzymes containing nonprimordial isotopes in cancer cells with no effect on normal cells. The patient’s tumor irradiated with thermal neutrons of specific kinetic energies to absorb under simultaneous conditions of RF and X-ray irradiations so as to enhance the selective absorbance of the neutrons by the nonprimordial isotopes to transmute the 13C to 14N, 15N to 16O, 17O to 19F for total inactivation of enzymes of glycolysis only in cancer cells with no affect on normal cells. In considering the merit of the theory 1, 2, 3 proposed for cancer cure, it is important to note that the radiofrequency is almost innocuous, but affects biomolecules in this newly discovered way. The radiowaves can be selected to only stop glycolysis in cancer with innocuous affects on normal cells. Soft X-rays can of certain wavelengths be innocuous to more biomolecules and tissue. X-rays can be tuned by specific wavelengths to excite only nonprimordial isotopes to selectively kill cancer cells with few effects on primordial isotopes and normal cells. Neutrons have no charge. They pass through most elements with no absorption. In this invention, tuning the neutrons by slowing and rotating the nonprimordial isotopes by modulated RF and specific wavelength of X-rays can selectively increase absorption cross-sections of the nonprimordials for neutron absorptions and transmutations. All three (radiowaves, X-rays and neutrons) can penetrate the whole human body for effective treatment of the whole body..
Conclusion
The analysis of DNA by MALDI mass spectroscopy led to the observations of different isotopic enrichments of nucleotides of guanosine (G), adenosine (A), cytidine (C), thymidine (T) and uridine (U). Such isotopic differences were further determined due to addition and removal of clumped isotopic enrichments of functional groups of 2D, 13C, 15N, 17O, and 25Mg associated with H, CH3, NH2, and OH. The DNA cancer cells tend to show enrichments with clustered kernels of 13CH3 relative to DNA of normal cells. The surrounding nanowaters and proteins were reasoned to accumulate 17O for hydrolysis to place 17O onto G, T, and C. The 17O was reasoned to lower activation for 13CH3 and 15NH2 functionalization of nucleotides. The accumulations of ammonia about cancer micro-environment were determined to facilitate such 15NH2 functionalizations. These novel nonprimordial functionalizations of purines and pyrimidines of nucleotides are consistent with observed fragmentations of DNA of cancer and normal cells.
On the basis of such totally new chemical dynamics as driven by nuclear magnetic moments (NMMs) of nonprimordial isotopes of 2H, 13C, 15N, 17O, and 25Mg of different NMMs cause ease of 17OH functionalizations and defunctionalizations with 17OH catalyzing 15NH2 and 13CH3 functionalizations. The resulting 13CH3 functionalizations cause difficult 13CH3 defuntionalizations for accumulations of U as T → T and → C but the 13CH3 defunctionalizations of T is kinetically hindered so T accumulates as U and C convert to T. Furthermore, the OH defunctionalizations of G to form A are accelerated but the functionalizations of A to G, which are kinetically hindered. Thereby with diet the host accumulates 2H, 13C, 15N, and 17O for altered functionalizations of U, T, C, A, and G; so that the clumped nonprimordial isotopes in the DNA cause internal chemical transformations of U → T and C → T and G → A with the developed deficiencies of G and C for causing the normal cell to transform to cancer cells.
On the basis of such accumulations of A and T in cells with deficiencies of G and C as discovered in this work by the model 1, 2, 3, many mutations are explained and model for cancer genesis. For instance, the lack of G and C by this model leads to the inability of RNA to properly translate some proteins like Cys, Trp and Arg. Such inability to translate these proteins correlates with the deficiency of Try, Arg and Cys in microcancer environments. Such alterations of protein translations on basis of functionalizations of nucleotides by nonprimordial isotopes provide a new mechanism for protein splicing for cancer genesis. The induced low translation of Try and Arg due to low G and C content further explains the unusual interactions of cancer with plasmodium malaria bacteria, which is known to have low G and C content in its DNA. Thereby, it is explained how bacteria of malaria may treat cancer. But other bacteria may cause cancer.
This model 1, 2, 3 by its determination of deficient G and C in cancer cells accounts for many mutations associated with cancer habitat. Less frequent cancer in whales, elephants, mole rats and bats can be reasoned by this low G and C due to nonprimordial isotopes as presented in this model. Low G and C has been determined to cause low melting point of DNA; such low G and C in cancer DNA relative to normal DNA is a basis for heat sensitivity of cancer cells. Weaker interactions by less G and C in cancer makes less rigid nucleus in cancer cells. By using the deficient G and C discovered in cancer in this research the altered cancer metabolism in zero gravity is understood in a new way as changing gravity would change the force fields about the cancer cells and alter its softer DNA relative to DNA of normal cells for selective killing cancer cells in zero gravity. Telomerase has the associate RNA with repeating sequences TTAGGG; so that low G and C would prevent RNA for expressing telomerase as is the character of cancer cell. So the low G and C in cancer DNA explains the less expressions of telomerase and the shortening of telomeres in cancer. The unusual G and C in bats can be explained by this model by the awkward flying and forces on bats for genetic mutations for higher contents of G and C with consequent unusual proteins in bats with explanations of ebola virus as it originates from bats. Whales and large mass and buoyancy without gravity for developing unusual G and C contents for long life of bats and whales. In general, fungi have high G and C content and the anti-cancer properties of fungi may be correlated to such for instance the unusual protein in brown seaweed fungi for forming polysaccharides in these seaweeds in environment rich in nonprimordial isotopes. The ease of mutation of C by 13CH3 methylation may explained how DNA was designed to involve stops that lack C. This theory 1, 2, 3 explains how and why the lack of G and C correlates with less Gly and Pro and altered formations of alpha helical blocker and induction of beta turns.
New theory 1, 2, 3 for eating animal DNA and cancer is given. By this new theory 1, 2, 3 it is further predicted that animals that eat animals have a greater possibility of cancer relative to herbivorous as the animal tissue have cells with DNA that enrich in nonprimordials and mutate the DNA in the animals that eat the DNA. But plant DNA has less nonprimordials. Eating animal DNA may cause cancer. Eating plant DNA may cure cancer. This theory 1, 2, 3 may explain why plants do not get cancer due to their lack of motion and their use of wind to move. But bats have huge motions for high levels of G and C in bat DNA; and plants are low G to C and bats high G to C. Therefore, by this theory the ability of animals and humans to move, crawl, walk, fly and swim causes needed extra catabolism with nonadiabatics (shaking nonprimordials for hidden dynamics of RBL) for greater incorporation of nonprimordials in animals and humans and cancer genesis. But trees lack such extensive catabolism and motions and accumulate less nonprimordials for less cancer in plants. For some reason grapes have seeds high in nonprimordials. Grape seeds have DNA that exchange nonprimordials with cancer DNA to kill the cancer relative to normal cells. Even greater eating of nonprimordials by host may lead to new technologies for treating and curing cancer by selectively stimulating the accumulated nonprimordial isotopes in the cancer cells.
References
- 1.Little R B. (2009) The Ferrochemistry of Carbon Nanotube, Diamond, Nucleic Acids and Proteins: The Magnetic Synergism of Macromolecules and Life’s Chemical Patterns. Carbon Nanotube: New Research. Ottenhouse A. , New York 223.
- 2.Little R B. (2018) On the atomic Carcinogenic mechanism and cure for cancer: Ferrochemistry for cause of Warburg Effect. , B P International 27.
- 4.Mehrez F, Bougatef K, Monache E D, Arisi I, de Santis LP et al. (2019) Telomere length measurement in tumor and on-tumor cells as a valuable prognostic for tumor progression. , Cancer Genetics 238, 50-61.
- 5.Uziel O. (2019) Cancer Cells Possess Different Isotopic Enrichment.https://doi.org/10.26434/chemrxiv.9989711.v1.
- 7.Zhang M, Sun J, Chen P. (2017) Chemical structure of proanthocyanidins. A Computational Tool for Accelerated Analysis of Oligomeric Proanthocyanidins in Plants. , J Food Compost Anal.; 56, 124-133.
- 8.Kawahara S, Ishihara S C, Matsumoto K, Senga S, Kawaguchi K et al. (2019) Identification and characterization of oligomeric proanthocyanidins (PAC) with significant anti-cancer activity in adzuki beans (Vigna angularis). , Heliyon 5, 02610.
- 9. (2014) Figure 4 in this Manuscript taken from; The Amino Acis Specified by Each mRNA Codon. Multiple Codons Can Code for the Same Amino Acids. The Information. in DNA via Translation. https://www.nature.com/scitable/topicpage/the-information-in-dna-determines-cellular-function-6523228/. © 2014 Nature Education .
- 10.M, Bird A. (2008) DNA methylation landscape: provocative insights from epigenomics. , Nat. Rev. Genet 9, 465.
- 11.Buchachenko A L, Kuznetsov D A, Berdinski V L. (2006) New Mechanism of Biological Effects of Electromagnetic Fields. , Biophysics 51, 489-496.
- 12.Chu C, Liu B, Plangger R, Kreutz C, Al-Hashimi H M. (2019) . M6A Minimally Impacts the Structure, Dynamics, and Rev ARM Binding Properties of HIV-1 RRE Stem IIB. doi: https://doi.org/10.1101/817940 .
- 13.T J Robinson, J A Freedman, M A Abo, A E Deveaux, LaCroix B et al. (2019) Alternative RNA Splicing as a Potential Major or Source of Untapped Molecular Targets in Precision Oncology and Cancer Disparities. American Association fo cancer Research. , DOI: 10-1158.
- 14.Garrett-Bakelman F E, Darshi M, Green S J, Gur R C, Lin L et al. (2019) The NASA Twins Study: A multidimensional Analysis of a Year Long Human Spaceflight. , Science 364, 144.
- 15.Madison R S, Hu X, Ramanan V, Xu Z, RSP Huang et al. (2022) Clustered 8-oxo-guanine mutations and oncogenic gene fusions in microsatellite-unstable colorectal cancer. , JCO Precision Oncology; 6, 2100477.
- 16.ML Van den Boogaard, Oka R, Hakkert A, Schold L, Ebus M E et al. (2022) Defects in 8-oxo-guanine repair pathways cause high frequency of C>A substitutions in neuroblastoma. PNAS. 118-36.
- 17.Krishnan R, Murugia M, Padma Lakshmi N, Mahalingam S. (2022) Guanine nucleotide binding protein like-1(GNC1) promotes cancer cell proliferation and survival through AKT/p21(CIP1) signaling cascade. , Molecular Biology of the Cell 31, 26.
- 18.Fang H, Zhu X, Yang H, J O H, Barbour J A et al. (2021) Deficiency of replication – independent DNA mismatch repair drives a 5-methycytosine deamination mutational signature in cancer. , Science Advances 7(45).
- 19.Prasad R, Yen T J, Bellacosa A. (2021) Active DNA depmethylation – The epigenetic gate keeper of development, immunity and cancer. Advanced Genetics. 2-1.
- 20.Sawicka M, Sawicka K, Lyson T, Politynska B, Miltyk W. (2022) Proline metabolism in malignant gliomas: a systematic literature review. Cancers. 14.
- 21.Firdous S, Abid R, Nawaz Z, Bukhari F, Anwer A et al. (2021) Dysregulated alanine as a potential predictive marker of glioma – An insight from untargeted HRMAS-NMR ad machine learning data. Metabolites. 11, 507.
- 22.Tajan M, Hennequart M, Cheung E C, Zani F, Hock A K. (2021) Serine synthesis pathway inhibition cooperates with dietary serine and glycine limitation for cancer therapy. , Nature Communications 12-366.
- 24.Chen C L, Hsu S C, Chung T Y, Chu C Y, Wang H J et al.Arginine is an epigenetic regulator targeting TEAD4 to modulate OXPHOS in prostate cancer cells. , Nature Communications(2021); 12, 2398.
- 25.M R Sullivan, L V Danai, C A Lewis, S H Chan, D Y Gui et al. (2019) Quantification of microenvironment metabolites in murine cancers reveal determinants of tumor nutrient availability. eLife. 8:e44235 DOI: 10.7554/eLife.44235 .
- 26.A B Howell, J D Reed, C G Krueger, Winterbottom R, D G Cunningham et al. (2005) Grape Seed Proanthocyanidins Structures; A-type cranberry proanthocyanidins and uropathogenic bacterial anti-adhesion activity. , Phytochemistry 66, 2281-22.
Cited by (1)
This article has been cited by 1 scholarly work according to:
Citing Articles:
Reginald B. Little - European Journal of Applied Physics (2023) Semantic Scholar

